CopciAB_103773
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_103773 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 103773 |
| Uniprot id | Functional description | Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin- like GTPase superfamily. Septin GTPase family | |
| Location | scaffold_7:1819921..1823430 | Strand | - |
| Gene length (nt) | 3510 | Transcript length (nt) | 3008 |
| CDS length (nt) | 2469 | Protein length (aa) | 822 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB17826 | 61 | 1.716E-304 | 976 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1113559 | 56.7 | 5.256E-278 | 867 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1456224 | 56.5 | 1.739E-274 | 857 |
| Pleurotus ostreatus PC9 | PleosPC9_1_57917 | 58.4 | 1.447E-274 | 857 |
| Agrocybe aegerita | Agrae_CAA7261592 | 58.5 | 2.479E-270 | 845 |
| Lentinula edodes NBRC 111202 | Lenedo1_1093431 | 53.1 | 2.706E-248 | 781 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_2248 | 53.1 | 2.686E-248 | 781 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_39848 | 52.9 | 3.839E-240 | 757 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_179274 | 46.7 | 4.619E-237 | 748 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_151367 | 47.4 | 6.8E-213 | 678 |
| Flammulina velutipes | Flave_chr05AA00201 | 49 | 3.328E-212 | 676 |
| Schizophyllum commune H4-8 | Schco3_2197600 | 41.5 | 1.035E-173 | 564 |
| Lentinula edodes B17 | Lened_B_1_1_13206 | 51.7 | 1.527E-160 | 525 |
| Auricularia subglabra | Aurde3_1_1272025 | 30.5 | 1.126E-102 | 355 |
| Grifola frondosa | Grifr_OBZ71217 | 57.7 | 2.895E-89 | 314 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_103773.T0 |
| Description | Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin- like GTPase superfamily. Septin GTPase family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd00067 | GAL4 | IPR001138 | 482 | 516 |
| CDD | cd00067 | GAL4 | IPR001138 | 707 | 737 |
| Pfam | PF00735 | Septin | IPR030379 | 31 | 161 |
| Pfam | PF00735 | Septin | IPR030379 | 207 | 272 |
| Pfam | PF00735 | Septin | IPR030379 | 354 | 406 |
| Pfam | PF00172 | Fungal Zn(2)-Cys(6) binuclear cluster domain | IPR001138 | 486 | 523 |
| Pfam | PF00172 | Fungal Zn(2)-Cys(6) binuclear cluster domain | IPR001138 | 707 | 741 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001138 | Zn(2)Cys(6) fungal-type DNA-binding domain |
| IPR030379 | Septin-type guanine nucleotide-binding (G) domain |
| IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| IPR036864 | Zn(2)-C6 fungal-type DNA-binding domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | MF |
| GO:0006355 | regulation of transcription, DNA-templated | BP |
| GO:0008270 | zinc ion binding | MF |
| GO:0005525 | GTP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| D | Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin- like GTPase superfamily. Septin GTPase family |
| U | Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin- like GTPase superfamily. Septin GTPase family |
| Z | Belongs to the TRAFAC class TrmE-Era-EngA-EngB-Septin- like GTPase superfamily. Septin GTPase family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| c6 zn |
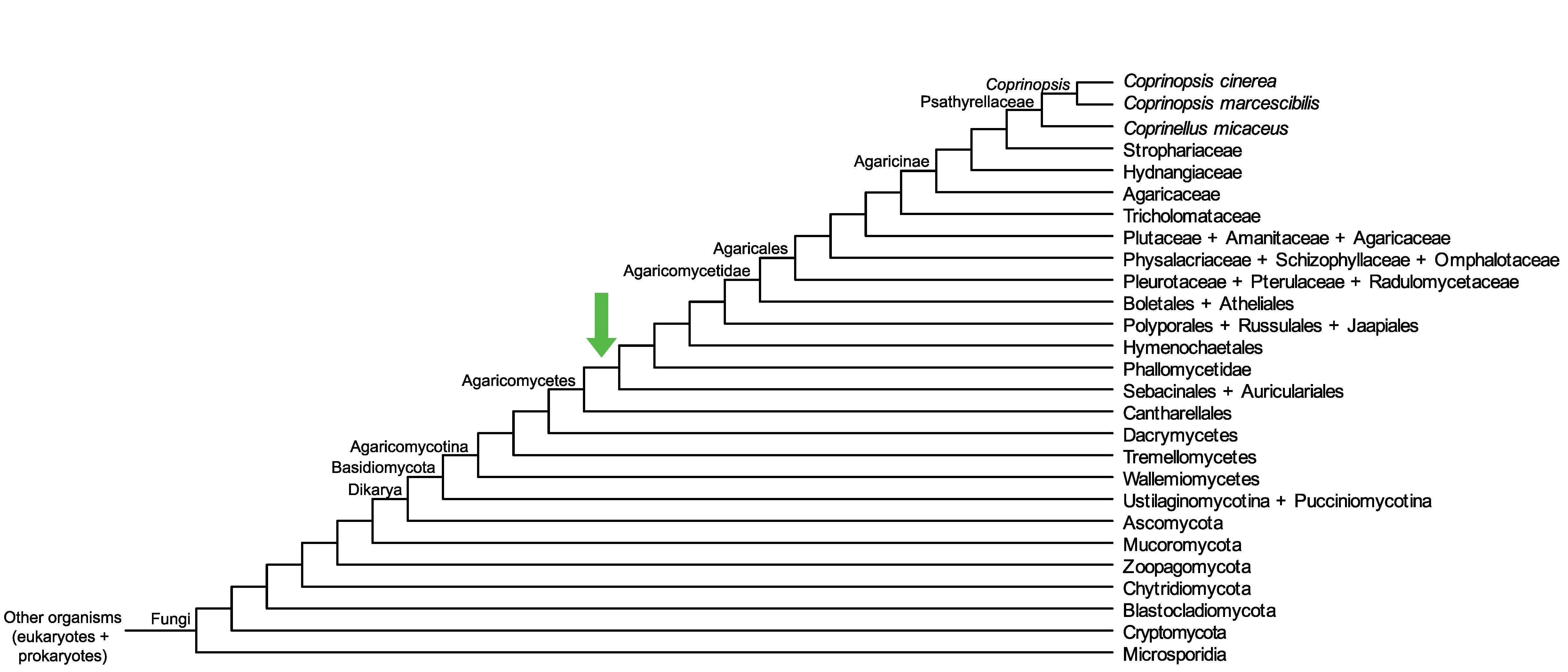
Conservation of CopciAB_103773 across fungi.
Arrow shows the origin of gene family containing CopciAB_103773.
Protein
| Sequence id | CopciAB_103773.T0 |
|---|---|
| Sequence |
>CopciAB_103773.T0 MNPRDHRIRHDDRDPNAQPNAYEAPDTARSEFTLLVAGCRGGKTSFLRLLLDTSDVSPTTTKDQLASVAKFVQGC ACHTTHIRTASLDIEIADGPNTRRPLGLNLIDTPSFDFHDEQEAERLMADVLHHIDSRFVEGLEDEWKARSGDRY VHLCIYFLDPDEIVPPTALGPPAPVVPRARNGSFSQPEEPVILEPPVNNPYMLRPVLPKADIAAIRRLSGRVNVL PVIARADVLSNERLAAVKVAIRRDLAEAGIGFGIFDMDGQYQQPQEDRSESSNGYGGYPNGTSTSNGNSPPVSPV TPAALRLPYALISPDAYSHSDGVPRKVPSRHELMQQYAAQFAHASKLARGKFVRTYRWGILDVLDPTHSDFLSLR SAIFHHMETLQKYTKEYLFSKFRQEVQLQHQKRPPSHHSVSQMSMLPSNSRPILAIDTAPQAIHRQPPPLHHGEM HPTSSARPMQDSAVGSGSAKSSTKTNRQRTKKITVACNFCRSRKLKCDGGRPACGQCTKRNNPCDYMPQNKRRGT VSKTSQRPKGEESESDSGDEGRSPEPSVSPQVSSQTVSRRSSNVDKLHPDGYHPPNTLPPLNDRRDVYGSGVAGS SSVSVNGLSASVPLPGPPSGPSSRAKILHPPEPPQHNYYNEKNELPHIATLLPDASPSTPAPMSAPSLSLAPIRP ASEQQAALRKRAATVPSKTTRLAGTSGPKVVACNFCRARKTKCDGAHPACASCARRQLACNYAHDSTASGLKKRR ASTSKTTADSPESLSPPSSRMAPTPSIGTDMSREAGDHLDDELELKRPMDHSEFGRVTKKMRMDDSGVSIAQ |
| Length | 822 |
Coding
| Sequence id | CopciAB_103773.T0 |
|---|---|
| Sequence |
>CopciAB_103773.T0 ATGAACCCAAGAGACCATCGCATAAGGCATGACGACCGAGACCCCAACGCTCAGCCCAACGCCTATGAGGCCCCA GACACCGCCCGCTCAGAGTTCACCCTTCTCGTAGCTGGCTGTCGAGGAGGCAAGACCTCATTTCTCCGCCTCCTG CTCGACACCAGCGACGTCTCCCCAACGACAACAAAGGACCAGCTCGCCTCCGTCGCAAAGTTCGTCCAGGGCTGC GCATGCCACACAACACATATCAGAACCGCGTCCCTCGACATAGAAATCGCGGATGGTCCCAACACAAGGCGACCC CTAGGTCTCAATCTCATCGACACACCCTCCTTTGATTTCCACGACGAGCAGGAAGCTGAACGCCTGATGGCCGAC GTCCTCCATCATATCGACTCGCGTTTTGTTGAAGGTCTTGAAGATGAGTGGAAGGCGCGCAGTGGCGACCGATAT GTGCATCTATGCATCTACTTCCTGGACCCGGATGAAATCGTGCCCCCTACGGCATTGGGTCCCCCTGCTCCGGTG GTTCCACGAGCTCGGAATGGTTCTTTCTCCCAGCCCGAAGAACCCGTTATCCTCGAGCCTCCGGTCAATAACCCG TACATGCTTCGGCCTGTCCTTCCCAAGGCTGACATTGCGGCCATTCGCCGTCTGTCGGGACGAGTCAACGTCCTG CCTGTCATTGCCAGGGCGGATGTGCTCAGCAACGAACGCCTCGCCGCTGTTAAGGTGGCTATCCGGAGGGACCTG GCGGAAGCGGGCATCGGGTTCGGGATATTCGACATGGACGGGCAGTACCAGCAACCCCAGGAGGACAGGTCGGAG TCTTCCAATGGCTATGGTGGTTACCCCAACGGAACATCAACATCCAACGGCAATTCTCCCCCGGTCTCCCCTGTA ACGCCTGCTGCACTGCGTTTGCCTTACGCACTCATCTCCCCTGACGCTTACTCCCACAGCGATGGTGTACCCCGA AAGGTTCCATCACGTCACGAACTCATGCAGCAGTATGCGGCTCAGTTCGCACATGCCTCCAAACTAGCCCGGGGC AAATTCGTTCGAACATATCGATGGGGGATTCTGGACGTACTCGACCCGACTCACAGCGACTTCCTTTCACTCCGA TCTGCTATATTCCATCATATGGAGACTTTACAGAAATACACGAAGGAGTATCTCTTCTCGAAGTTTCGACAAGAA GTTCAGCTGCAGCACCAGAAACGACCCCCTTCGCACCACTCTGTTTCACAAATGTCCATGCTTCCTTCAAATTCG CGGCCCATTTTGGCTATTGATACCGCACCCCAGGCGATTCATCGCCAACCGCCACCCCTCCATCATGGTGAAATG CACCCCACTTCGTCGGCGCGCCCCATGCAGGACAGCGCGGTGGGAAGTGGTTCTGCCAAGTCTTCGACAAAGACG AACAGGCAAAGAACGAAAAAGATTACGGTTGCCTGCAATTTCTGCCGATCTCGCAAATTGAAATGTGATGGGGGA CGCCCTGCATGTGGCCAGTGCACGAAGCGGAACAATCCCTGTGACTATATGCCTCAGAACAAACGGCGTGGAACG GTCAGTAAGACCTCACAGCGTCCGAAAGGTGAGGAGAGTGAGAGTGACAGTGGTGATGAGGGTCGCTCCCCTGAA CCCTCTGTATCACCTCAAGTTTCGTCACAGACTGTGTCTCGCCGCAGTTCAAATGTGGACAAACTTCACCCCGAT GGTTATCATCCTCCCAACACACTCCCACCATTGAATGATCGTCGGGATGTCTACGGAAGCGGTGTCGCCGGTTCC TCGTCCGTGAGCGTTAACGGGCTCTCTGCTTCTGTTCCGCTCCCTGGGCCTCCTTCCGGACCCAGTTCGCGGGCA AAGATCCTCCATCCCCCCGAACCTCCGCAGCACAACTATTACAACGAAAAGAACGAGTTACCTCACATCGCGACT TTGCTGCCCGACGCTTCGCCTTCGACTCCTGCGCCAATGTCGGCGCCCTCACTCTCCCTTGCTCCCATTCGACCC GCCTCTGAGCAGCAGGCTGCGTTACGGAAGCGCGCGGCTACTGTCCCCAGTAAGACTACTCGACTCGCTGGAACC TCTGGGCCCAAAGTAGTAGCATGCAACTTTTGTCGAGCTCGTAAAACCAAGTGCGATGGGGCACACCCTGCATGT GCAAGCTGCGCTCGCAGGCAACTTGCTTGCAACTATGCCCACGACAGCACGGCGTCTGGACTGAAGAAAAGACGA GCTTCGACCTCGAAAACGACCGCCGACTCTCCCGAGTCGCTTTCTCCACCCTCCTCGCGGATGGCGCCTACCCCG TCGATCGGGACCGACATGTCTCGCGAGGCCGGTGATCATCTCGACGATGAACTAGAATTGAAACGACCCATGGAC CACTCGGAATTTGGCCGAGTGACGAAGAAAATGAGGATGGACGACAGTGGGGTCTCCATCGCTCAGTAA |
| Length | 2469 |
Transcript
| Sequence id | CopciAB_103773.T0 |
|---|---|
| Sequence |
>CopciAB_103773.T0 ATCTTTCAGACTGGACCTTCTTCCGTCCAGCGAAGGCTGCCCAGAAAGGCCCTGCTCGTCGTCCAAAACAGCCTC CAGCGTCCATGAACCCAAGAGACCATCGCATAAGGCATGACGACCGAGACCCCAACGCTCAGCCCAACGCCTATG AGGCCCCAGACACCGCCCGCTCAGAGTTCACCCTTCTCGTAGCTGGCTGTCGAGGAGGCAAGACCTCATTTCTCC GCCTCCTGCTCGACACCAGCGACGTCTCCCCAACGACAACAAAGGACCAGCTCGCCTCCGTCGCAAAGTTCGTCC AGGGCTGCGCATGCCACACAACACATATCAGAACCGCGTCCCTCGACATAGAAATCGCGGATGGTCCCAACACAA GGCGACCCCTAGGTCTCAATCTCATCGACACACCCTCCTTTGATTTCCACGACGAGCAGGAAGCTGAACGCCTGA TGGCCGACGTCCTCCATCATATCGACTCGCGTTTTGTTGAAGGTCTTGAAGATGAGTGGAAGGCGCGCAGTGGCG ACCGATATGTGCATCTATGCATCTACTTCCTGGACCCGGATGAAATCGTGCCCCCTACGGCATTGGGTCCCCCTG CTCCGGTGGTTCCACGAGCTCGGAATGGTTCTTTCTCCCAGCCCGAAGAACCCGTTATCCTCGAGCCTCCGGTCA ATAACCCGTACATGCTTCGGCCTGTCCTTCCCAAGGCTGACATTGCGGCCATTCGCCGTCTGTCGGGACGAGTCA ACGTCCTGCCTGTCATTGCCAGGGCGGATGTGCTCAGCAACGAACGCCTCGCCGCTGTTAAGGTGGCTATCCGGA GGGACCTGGCGGAAGCGGGCATCGGGTTCGGGATATTCGACATGGACGGGCAGTACCAGCAACCCCAGGAGGACA GGTCGGAGTCTTCCAATGGCTATGGTGGTTACCCCAACGGAACATCAACATCCAACGGCAATTCTCCCCCGGTCT CCCCTGTAACGCCTGCTGCACTGCGTTTGCCTTACGCACTCATCTCCCCTGACGCTTACTCCCACAGCGATGGTG TACCCCGAAAGGTTCCATCACGTCACGAACTCATGCAGCAGTATGCGGCTCAGTTCGCACATGCCTCCAAACTAG CCCGGGGCAAATTCGTTCGAACATATCGATGGGGGATTCTGGACGTACTCGACCCGACTCACAGCGACTTCCTTT CACTCCGATCTGCTATATTCCATCATATGGAGACTTTACAGAAATACACGAAGGAGTATCTCTTCTCGAAGTTTC GACAAGAAGTTCAGCTGCAGCACCAGAAACGACCCCCTTCGCACCACTCTGTTTCACAAATGTCCATGCTTCCTT CAAATTCGCGGCCCATTTTGGCTATTGATACCGCACCCCAGGCGATTCATCGCCAACCGCCACCCCTCCATCATG GTGAAATGCACCCCACTTCGTCGGCGCGCCCCATGCAGGACAGCGCGGTGGGAAGTGGTTCTGCCAAGTCTTCGA CAAAGACGAACAGGCAAAGAACGAAAAAGATTACGGTTGCCTGCAATTTCTGCCGATCTCGCAAATTGAAATGTG ATGGGGGACGCCCTGCATGTGGCCAGTGCACGAAGCGGAACAATCCCTGTGACTATATGCCTCAGAACAAACGGC GTGGAACGGTCAGTAAGACCTCACAGCGTCCGAAAGGTGAGGAGAGTGAGAGTGACAGTGGTGATGAGGGTCGCT CCCCTGAACCCTCTGTATCACCTCAAGTTTCGTCACAGACTGTGTCTCGCCGCAGTTCAAATGTGGACAAACTTC ACCCCGATGGTTATCATCCTCCCAACACACTCCCACCATTGAATGATCGTCGGGATGTCTACGGAAGCGGTGTCG CCGGTTCCTCGTCCGTGAGCGTTAACGGGCTCTCTGCTTCTGTTCCGCTCCCTGGGCCTCCTTCCGGACCCAGTT CGCGGGCAAAGATCCTCCATCCCCCCGAACCTCCGCAGCACAACTATTACAACGAAAAGAACGAGTTACCTCACA TCGCGACTTTGCTGCCCGACGCTTCGCCTTCGACTCCTGCGCCAATGTCGGCGCCCTCACTCTCCCTTGCTCCCA TTCGACCCGCCTCTGAGCAGCAGGCTGCGTTACGGAAGCGCGCGGCTACTGTCCCCAGTAAGACTACTCGACTCG CTGGAACCTCTGGGCCCAAAGTAGTAGCATGCAACTTTTGTCGAGCTCGTAAAACCAAGTGCGATGGGGCACACC CTGCATGTGCAAGCTGCGCTCGCAGGCAACTTGCTTGCAACTATGCCCACGACAGCACGGCGTCTGGACTGAAGA AAAGACGAGCTTCGACCTCGAAAACGACCGCCGACTCTCCCGAGTCGCTTTCTCCACCCTCCTCGCGGATGGCGC CTACCCCGTCGATCGGGACCGACATGTCTCGCGAGGCCGGTGATCATCTCGACGATGAACTAGAATTGAAACGAC CCATGGACCACTCGGAATTTGGCCGAGTGACGAAGAAAATGAGGATGGACGACAGTGGGGTCTCCATCGCTCAGT AAGTTTGCTATACCCATGTTTGTCTGTTGTTCCTCCTCGTCTTCTTTCATTCATGTGTAGCTCTTGCCACTGTCC ATTTGTTGTTTCTCCACCCGTTGTAGACTACTCCCTTCCCTTCTTCTGTCTGTCTCGGCTTCCATTCATCCAACC CATCCAATCATTGACTCGTCTCTTTCTCATGCTCACGCAACCACTCTAACATGCTCTCTGCTTTTCCCTTCTGAC TTGTATATACTTTTCTTCATGATCATGAAACACCAATTTTCTTCTTTTGGCTCCTCCGCATTCTTGCGTCCGTCT GTTTTCTTTTTGGGCTTTTGGGCGTTAGTTAAATAGCTAGCTATCTTTTTCTTTTTTGCATATTGCAGCATGACT TTACTCTTCACGTTGTGTACTTGTTATTAGCGAAGAAATTGTTGTTACACTTGCATTTGATGAATCCATAGGGGA TGAACTGT |
| Length | 3008 |
Gene
| Sequence id | CopciAB_103773.T0 |
|---|---|
| Sequence |
>CopciAB_103773.T0 ATCTTTCAGACTGGACCTTCTTCCGTCCAGCGAAGGCTGCCCAGAAAGGCCCTGCTCGTCGTCCAAAACAGCCTC CAGCGTCCATGAACCCAAGAGACCATCGCATAAGGCATGACGACCGAGACCCCAACGCTCAGCCCAACGCCTATG AGGCCCCAGGTAAGCCCGCCATGCGCCATGGGCCACCGTCTCCGGGCCAGCCCTGCTCATCGTCCATCGCCGTTC CACAGACACCGCCCGCTCAGAGTTCACCCTTCTCGTAGCTGGCTGTCGAGGAGGTGAGTATTTTCTTCTTCTCGC CGTCCCGGAGTGCGCAATCCTCCAGAGGAACGTTCAATAACCCCCGTCGCTTTCGGATCACAACAGGCAAGACCT CATTTCTCCGCCTCCTGCTCGACACCAGCGACGTCTCCCCAACGACAACAAAGGACCAGCTCGCCTCCGTCGCAA AGTTCGTCCAGGGCTGCGCATGCCACACAACACATATCAGAACCGCGTCCCTCGACATAGAAATCGCGGATGGTC CCAACACAAGGCGACCCCTAGGTCTCAATCTCATCGACACACCCTCCTTTGATTTCCACGACGAGCAGGAAGCTG AACGCCTGATGGCCGACGTCCTCCATCATATCGACTCGCGTTTTGTTGAAGGTCTTGAAGATGTAAGCGCTAACT ATGTCATACCCATGCACCGCCCACACCATCAACTAATATCGCGCCCTTAGGAGTGGAAGGCGCGCAGTGGCGACC GATATGTGCATCTGTATGTCTCCCCACTCTCCTTCCCCAACTCACTTGTCAACTCAGCACCGCCTCCTGTTAGAT GCATCTACTTCCTGGACCCGGATGAAATCGTGCCCCCTACGGCATTGGGTCCCCCTGCTCCGGTGGTTCCACGAG CTCGGAATGGTTCTTTCTCCCAGCCCGAAGAACCCGTTATCCTCGAGCCTCCGGTCAATAACCCGTACATGCTTC GGCCTGTCCTTCCCAAGGCTGACATTGCGGCCATTCGCCGTCTGTCGGGACGAGTCAACGTCCTGCCTGTCATTG CCAGGGCGGATGTGCTCAGCAACGAACGCCTCGCCGCTGTTAAGGTGGCTATCCGGAGGGACCTGGCGGAAGCGG GCATCGGGTTCGGGATATTCGACATGGACGGGCAGTACCAGCAACCCCAGGAGGACAGGTCGGAGTCTTCCAATG GCTATGGTGGTTACCCCAACGGAACATCAACATCCAACGGCAATTCTCCCCCGGTCTCCCCTGTAACGCCTGCTG CACTGCGTTTGCCTTACGCACTCATCTCCCCTGACGCTTACTCCCACAGCGATGGTGTACCCCGAAAGGTTCCAT CACGTCACGAACTCATGCAGCAGTATGCGGCTCAGTTCGCACATGCCTCCAAACTAGCCCGGGGCAAATTCGTTC GAACATATCGATGGGGGATTCTGGACGTACTCGACCCGACTCACAGCGACTTCCTTTCACTCCGATCTGCTATAT TCCATCATATGGAGGCAAGCATATTCCTGTCTCACTCGGTCGTTATTCAAAACCTTCTTACGTTTCCATTTCACA AACAGACTTTACAGAAATACACGAAGGAGTATCTCTTCTCGAAGTTTCGACAAGAAGTTCAGCTGCAGCACCAGA AACGACCCCCTTCGCACCACTCTGTTTCACAAATGTCCATGCTTCCTTCAAATTCGCGGCCCATTTTGGCTATTG ATACCGCACCCCAGGCGATTCATCGCCAACCGCCACCCCTCCATCATGGTGAAATGCACCCCACTTCGTCGGCGC GCCCCATGCAGGACAGCGCGGTGGGAAGTGGTTCTGCCAAGTCTTCGACAAGTGGGTACACTCCTGTCGGTTGGA GCGCCTTCAGACTGACCCTTTAATTTAACAGAGACGAACAGGCAAAGAACGAAAAAGATTACGGTTGCCTGCAAT TTCTGCCGATGTAAATCCGTTTCTTTACTGTTTAAGCTGTTACTGACCATTCATCTGCAGCTCGCAAATTGAAAT GTGATGGGGGACGCCCTGCATGTGGCCAGTGCACGAAGCGGAACAATCCCTGTGACTATATGCCTCAGAACAAAC GGCGTGGAACGGTCAGTAAGACCTCACAGCGTCCGAAAGGTGAGGAGAGTGAGAGTGACAGTGGTGATGAGGGTC GCTCCCCTGAACCCTCTGTATCACCTCAAGTTTCGTCACAGACTGTGTCTCGCCGCAGTTCAAATGTGGACAAAC TTCACCCCGATGGTTATCATCCTCCCAACACACTCCCACCATTGAATGATCGTCGGGATGTCTACGGAAGCGGTG TCGCCGGTTCCTCGTCCGTGAGCGTTAACGGGCTCTCTGCTTCTGTTCCGCTCCCTGGGCCTCCTTCCGGACCCA GTTCGCGGGCAAAGATCCTCCATCCCCCCGAACCTCCGCAGCACAACTATTACAACGAAAAGAACGAGTTACCTC ACATCGCGACTTTGCTGCCCGACGCTTCGCCTTCGACTCCTGCGCCAATGTCGGCGCCCTCACTCTCCCTTGCTC CCATTCGACCCGCCTCTGAGCAGCAGGCTGCGTTACGGAAGCGCGCGGCTACTGTCCCCAGTAAGACTACTCGAC TCGCTGGAACCTCTGGGCCCAAAGTAGTAGCATGCAACTTTTGTCGAGGTAAGAAGTGGAAAGGTTTAGTAGGTT CTGGCTGACATTTGGTTTGCAGCTCGTAAAACCAAGTGCGATGGGGCACACCCTGCATGTGCAAGCTGCGCTCGC AGGCAACTTGCTTGCAACTATGCCCACGACAGCACGGCGTCTGGACTGAAGAAAAGACGAGCTTCGACCTCGAAA ACGACCGCCGACTCTCCCGAGTCGCTTTCTCCACCCTCCTCGCGGATGGCGCCTACCCCGTCGATCGGGACCGAC ATGTCTCGCGAGGCCGGTGATCATCTCGACGATGAACTAGAATTGAAACGACCCATGGACCACTCGGAATTTGGC CGAGTGACGAAGAAAATGAGGATGGACGACAGTGGGGTCTCCATCGCTCAGTAAGTTTGCTATACCCATGTTTGT CTGTTGTTCCTCCTCGTCTTCTTTCATTCATGTGTAGCTCTTGCCACTGTCCATTTGTTGTTTCTCCACCCGTTG TAGACTACTCCCTTCCCTTCTTCTGTCTGTCTCGGCTTCCATTCATCCAACCCATCCAATCATTGACTCGTCTCT TTCTCATGCTCACGCAACCACTCTAACATGCTCTCTGCTTTTCCCTTCTGACTTGTATATACTTTTCTTCATGAT CATGAAACACCAATTTTCTTCTTTTGGCTCCTCCGCATTCTTGCGTCCGTCTGTTTTCTTTTTGGGCTTTTGGGC GTTAGTTAAATAGCTAGCTATCTTTTTCTTTTTTGCATATTGCAGCATGACTTTACTCTTCACGTTGTGTACTTG TTATTAGCGAAGAAATTGTTGTTACACTTGCATTTGATGAATCCATAGGGGATGAACTGT |
| Length | 3510 |