CopciAB_108063
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_108063 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 108063 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_2:2217503..2220051 | Strand | - |
| Gene length (nt) | 2549 | Transcript length (nt) | 1694 |
| CDS length (nt) | 1566 | Protein length (aa) | 521 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21515 | 51 | 1.577E-174 | 552 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1446746 | 45.5 | 1.904E-150 | 482 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1099610 | 43.5 | 2.211E-146 | 470 |
| Agrocybe aegerita | Agrae_CAA7267978 | 46.6 | 3.67E-142 | 458 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_108151 | 35.6 | 5.881E-109 | 361 |
| Schizophyllum commune H4-8 | Schco3_2484255 | 42 | 1.218E-89 | 305 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_194189 | 49.4 | 4.67E-85 | 291 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_108063.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd21037 | MLKL_NTD | - | 28 | 161 |
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 260 | 516 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR036537 | Adaptor protein Cbl, N-terminal domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
| GO:0007166 | cell surface receptor signaling pathway | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
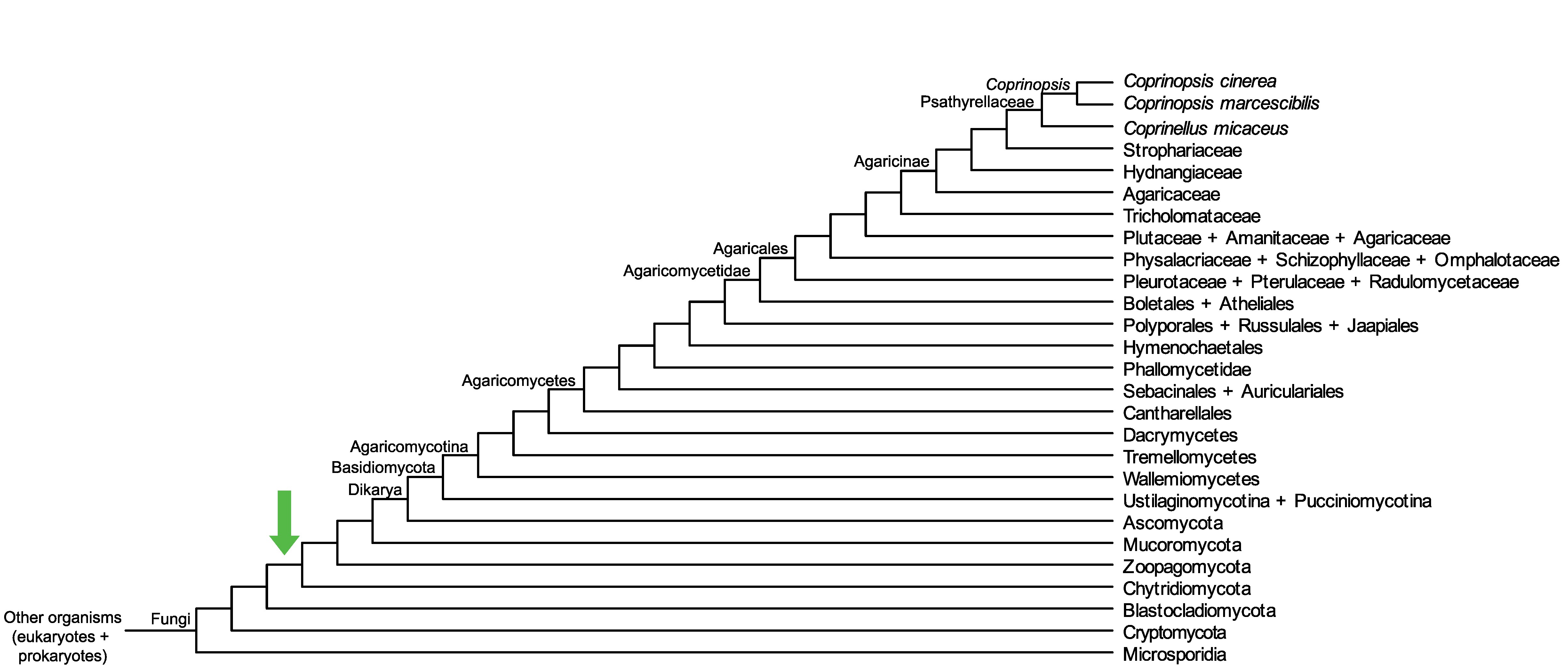
Conservation of CopciAB_108063 across fungi.
Arrow shows the origin of gene family containing CopciAB_108063.
Protein
| Sequence id | CopciAB_108063.T0 |
|---|---|
| Sequence |
>CopciAB_108063.T0 MMAKDLVSGILGAVELGANVAGVPFAAGAALILKEIVDTCDQLRIHKKKARTIRNKCVQFTLLLDDHSRTNIDAE LLKSIDETRAIFERVYSRMHRYSQMSAWSLFLKMSEIGEGLDECEANVQSAIEMFQFAAHAVGNRNQQEIKETMR VQHEEMRALLREVLTKPSEAREIIAMQRSGSQVAQQVMQAGQEQLRNELMISNTRSESPTREGRDQEATTQKRRP GSDIRDALYQFHLQTGTLPAIKILNEEITRRSDHAVAGGTHSDIWEGLWLGRKKVALKGLRNIKADDPRAKLRIE REIKTWSSLQNEHILPFYGIVTDIGTHIHMVSPWQENGNVLDYLKKQPNVNRLRIIKGAARGLAYLHSMKIVHGN VKCTNILVMDNGEACICDFGMSKLIEEVTEKAASATLTAAGSARWLAPELIDGTISSPTFATDTYSFAMAVLELL TGKYPFAERKRDASVIHDVVVMKQTPARPQDPRVCVWLTDELWNLLGGCWHNSAESRPTMAEVTGELDLFE |
| Length | 521 |
Coding
| Sequence id | CopciAB_108063.T0 |
|---|---|
| Sequence |
>CopciAB_108063.T0 ATGATGGCTAAGGACCTCGTCTCTGGAATACTTGGTGCAGTCGAACTTGGGGCTAACGTCGCTGGAGTTCCATTC GCTGCCGGAGCCGCACTTATACTCAAAGAGATTGTCGATACTTGTGACCAGCTTCGTATTCACAAGAAGAAGGCT AGAACTATCAGAAACAAATGCGTCCAGTTCACACTCCTGCTCGATGACCACTCCCGGACCAACATCGACGCCGAA CTGCTAAAGTCAATCGATGAAACCAGGGCGATCTTTGAACGAGTGTACTCCCGGATGCATAGGTATTCACAAATG AGCGCCTGGTCTCTATTCCTCAAGATGTCCGAGATCGGGGAAGGTCTGGACGAATGCGAGGCGAACGTTCAATCT GCCATTGAGATGTTCCAGTTCGCCGCCCATGCCGTAGGAAATAGAAACCAGCAAGAAATCAAAGAAACTATGCGG GTCCAGCATGAAGAAATGAGGGCCCTCCTCCGCGAGGTCCTCACTAAGCCCTCGGAGGCAAGGGAGATCATCGCA ATGCAGAGGTCAGGGAGCCAAGTCGCACAGCAAGTGATGCAAGCCGGTCAAGAGCAATTGCGCAATGAATTGATG ATCAGCAACACTCGATCCGAGTCCCCGACAAGAGAAGGAAGGGACCAGGAGGCCACAACCCAGAAAAGGCGCCCT GGAAGTGACATCCGGGATGCACTTTACCAATTCCACTTGCAAACTGGTACACTACCAGCCATAAAGATCCTCAAC GAGGAGATCACTAGACGCTCAGATCATGCAGTGGCAGGAGGAACTCACAGTGACATCTGGGAAGGGCTCTGGCTA GGCCGCAAAAAGGTCGCGCTCAAGGGCCTTAGGAACATAAAGGCTGATGATCCCAGGGCCAAGCTGCGAATCGAA CGCGAGATAAAGACGTGGTCATCGCTCCAGAACGAGCACATATTACCATTCTATGGGATCGTGACCGATATCGGG ACACATATACATATGGTTTCTCCATGGCAGGAGAACGGAAACGTTCTAGACTATCTCAAGAAACAACCCAACGTA AACCGTTTACGCATCATTAAAGGTGCGGCACGAGGCTTAGCCTACCTACACTCAATGAAGATAGTTCACGGAAAT GTGAAATGCACGAACATCCTTGTAATGGACAACGGGGAGGCGTGTATATGTGATTTTGGGATGTCCAAGCTGATC GAGGAGGTAACAGAGAAGGCAGCCTCGGCGACGTTGACGGCTGCAGGGAGTGCTCGCTGGTTAGCGCCGGAGCTG ATTGATGGGACCATTTCATCCCCGACGTTTGCGACGGACACCTACTCCTTCGCCATGGCGGTTCTGGAGTTACTT ACAGGGAAATACCCATTCGCAGAACGTAAGAGGGATGCGTCTGTAATACACGATGTGGTGGTCATGAAGCAGACG CCTGCTCGCCCGCAAGATCCGAGGGTTTGTGTGTGGTTGACGGATGAGCTGTGGAACCTCTTGGGAGGGTGCTGG CACAACTCGGCAGAATCGAGACCCACAATGGCGGAAGTGACAGGAGAATTGGATTTATTCGAGTAG |
| Length | 1566 |
Transcript
| Sequence id | CopciAB_108063.T0 |
|---|---|
| Sequence |
>CopciAB_108063.T0 GCATTGCTGTAGACTCCACCTCGCCGTTGTCGTTCCTTGTCCTCTGCCCCCTCATACATTCTATGATGGCTAAGG ACCTCGTCTCTGGAATACTTGGTGCAGTCGAACTTGGGGCTAACGTCGCTGGAGTTCCATTCGCTGCCGGAGCCG CACTTATACTCAAAGAGATTGTCGATACTTGTGACCAGCTTCGTATTCACAAGAAGAAGGCTAGAACTATCAGAA ACAAATGCGTCCAGTTCACACTCCTGCTCGATGACCACTCCCGGACCAACATCGACGCCGAACTGCTAAAGTCAA TCGATGAAACCAGGGCGATCTTTGAACGAGTGTACTCCCGGATGCATAGGTATTCACAAATGAGCGCCTGGTCTC TATTCCTCAAGATGTCCGAGATCGGGGAAGGTCTGGACGAATGCGAGGCGAACGTTCAATCTGCCATTGAGATGT TCCAGTTCGCCGCCCATGCCGTAGGAAATAGAAACCAGCAAGAAATCAAAGAAACTATGCGGGTCCAGCATGAAG AAATGAGGGCCCTCCTCCGCGAGGTCCTCACTAAGCCCTCGGAGGCAAGGGAGATCATCGCAATGCAGAGGTCAG GGAGCCAAGTCGCACAGCAAGTGATGCAAGCCGGTCAAGAGCAATTGCGCAATGAATTGATGATCAGCAACACTC GATCCGAGTCCCCGACAAGAGAAGGAAGGGACCAGGAGGCCACAACCCAGAAAAGGCGCCCTGGAAGTGACATCC GGGATGCACTTTACCAATTCCACTTGCAAACTGGTACACTACCAGCCATAAAGATCCTCAACGAGGAGATCACTA GACGCTCAGATCATGCAGTGGCAGGAGGAACTCACAGTGACATCTGGGAAGGGCTCTGGCTAGGCCGCAAAAAGG TCGCGCTCAAGGGCCTTAGGAACATAAAGGCTGATGATCCCAGGGCCAAGCTGCGAATCGAACGCGAGATAAAGA CGTGGTCATCGCTCCAGAACGAGCACATATTACCATTCTATGGGATCGTGACCGATATCGGGACACATATACATA TGGTTTCTCCATGGCAGGAGAACGGAAACGTTCTAGACTATCTCAAGAAACAACCCAACGTAAACCGTTTACGCA TCATTAAAGGTGCGGCACGAGGCTTAGCCTACCTACACTCAATGAAGATAGTTCACGGAAATGTGAAATGCACGA ACATCCTTGTAATGGACAACGGGGAGGCGTGTATATGTGATTTTGGGATGTCCAAGCTGATCGAGGAGGTAACAG AGAAGGCAGCCTCGGCGACGTTGACGGCTGCAGGGAGTGCTCGCTGGTTAGCGCCGGAGCTGATTGATGGGACCA TTTCATCCCCGACGTTTGCGACGGACACCTACTCCTTCGCCATGGCGGTTCTGGAGTTACTTACAGGGAAATACC CATTCGCAGAACGTAAGAGGGATGCGTCTGTAATACACGATGTGGTGGTCATGAAGCAGACGCCTGCTCGCCCGC AAGATCCGAGGGTTTGTGTGTGGTTGACGGATGAGCTGTGGAACCTCTTGGGAGGGTGCTGGCACAACTCGGCAG AATCGAGACCCACAATGGCGGAAGTGACAGGAGAATTGGATTTATTCGAGTAGTGATGTATGTATTTTGTAGTAG GTGTCTCCTGGCATTGACTCAACAACGACGACGGGGTTAACGAG |
| Length | 1694 |
Gene
| Sequence id | CopciAB_108063.T0 |
|---|---|
| Sequence |
>CopciAB_108063.T0 GCATTGCTGTAGACTCCACCTCGCCGTTGTCGTTCCTTGTCCTCTGCCCCCTCATACATTCTATGATGGCTAAGG ACCTCGTCTCTGGAATACTTGGTGCAGTCGAACTTGGGGCTAACGTCGCTGGAGTTCCATTCGCTGCCGGAGCCG CACTTATACTCAAAGAGATTGTCGATACTTGTGACCAGCTTCGTATTCACAAGGTGAGTGAAACATACAGCTTGA CCTCTGTATCTGCTTACATTGACTGTAGAAGAAGGCTAGAACTATCAGAAACAAATGCGTCCAGTTCACACTCCT GCTCGATGACCACTCCCGGACCAACATCGACGCCGAACTGCTAAAGTCAATCGATGAAACCAGGGCGTCAGTTCC TTTCCATTAATAGACATGGAATTACTCTCTTATGCTTCTTTCATTAGGATCTTTGAACGAGTGTACTCCCGGATG CATAGGTATTCACAAATGAGCGCCTGGTCTCTATTCCGCAAGTCATGCTTCCCCGTCACCCTATCACATTCCCTT TCTTACGCACAATCTCAGTCAAGATGTCCGAGATCGGGGAAGGTCTGGACGAATGCGAGGCGAACGTTCAATCTG CCATTGAGATGTTCCAGGTGGGTGCTTTATCCGTGCCACTTAGTGAGGTAACAACTTATCGAGTGGGACAGTTCG CGTAAGGCCCTGCTCTGCCTCTTGGCCCATGTCCCAATTCATTCACAGACACCTTCAGCGCCCATGCCGTAGGAA ATAGAAACCAGCAAGAAATCAAAGAAACTATGCGGGTCCAGCATGAAGAAATGAGGGCCCTCCTCCGCGAGGTCC TCACTAAGCCCTCGGAGGCAAGGGAGATCATCGCAATGCAGAGGTCAGGGAGCCAAGTCGCACAGCAAGTGATGC AAGCCGGTCAAGAGGTCTGCTTTCTCCTTGTTCAGTCTCTTCACCTACGTCATTGATATTTCCTTGCGGGCATAG CAATTGCGCAATGAATTGATGATCAGCAACACTCGATCCGAGTCCCCGACAAGAGAAGGAAGGGACCAGGAGGCC ACAACCCAGAAAAGGCGCCCTGGAAGTGACATCCGGGATGCACTTTACCAATTCCACTTGCAAACTGGTACACTA CCAGCCATAAAGATCCTCAACGAGGAGATCACTAGACGCTCAGATCATGCAGTGGCAGGAGGAACTCACAGTGAC ATCTGGGAAGGGCTCTGGCTAGGCCGCAAAAAGGTACTTCAAGCGAGTAGGGTATCCTATAATCTTTCAGAAACT CACGGACCCTGTTTCACAGGTCGCGCTCAAGGGCCTTAGGAACATAAAGGCTGATGATCCCAGGGCCAAGCTGGC AAGTTGACGATCGATTGGACACCTAACCTTCCGTTCTGACTGGGGAATCCAAAAACAGCGAATCGAACGCGAGAT AAAGACGTGGTCATCGCTCCAGAACGAGCACATATTACCATTCTATGGGATCGTGACCGATATCGGGACACATAT ACATATGGTATGTCATGGAAAGAACCACCCTAAAGACATTGGCTAAACATATCAGAAAGGTTTCTCCATGGCAGG AGAACGGAAACGTTCTAGAGTAGGCTTCATATGTCATACAAGCGTCACAAAGGGTTGGTGAACTGACACCTGCCC AAGCTATCTCAAGAAACAACCCAACGTAAACCGTTTACGCATCGTGAGTGGCCACGTCCCTGTCCTCGTTCCCTC CTATCGGGCTTACCTGTTTCTCTTCAGATTAAAGGTGCGGCACGAGGCTTAGCCTACCTACACTCAATGAAGATA GTTCACGGAAATGGTGCGACGATTTCTAATTTTACACCGATAAACTTACATGTGGATTGATGATCGATTCGTGGC TGGTGCGGTGTGGTTCCACCAGTGAAATGCGTACGTGATTCTTTCCCTTATTGGCCTTGTTTGAAACGTTCAGCT CACAAAGGTGTGAATGAAACTCTCCTCATACAGACGAACATCCTTGTAATGGACAACGTAAGGACGGGATTCTTT GAGGTTCTTCGGAACTTGGCGCTGACTATCAGGATGGGTCTCACGTACAGGGGGAGGCGTGTATATGTGATTTTG GGATGTCCAAGCTGATCGAGGAGGTAACAGAGAAGGCAGCCTCGGCGACGTTGACGGCTGCAGGGAGTGCTCGCT GGTTAGCGCCGGAGCTGATTGATGGGACCATTTCATCCCCGACGTTTGCGACGGACACCTACTCCTTCGCCATGG CGGTTCTGGAGTTACTTACAGGGAAATACCCATTCGCAGAACGTAAGAGGGATGCGTCTGTAATACACGATGTGG TGGTCATGAAGCAGACGCCTGCTCGCCCGCAAGATCCGAGGGTTTGTGTGTGGTTGACGGATGAGCTGTGGAACC TCTTGGGAGGGTGCTGGCACAACTCGGCAGAATCGAGACCCACAATGGCGGAAGTGACAGGAGAATTGGATTTAT TCGAGTAGTGATGTATGTATTTTGTAGTAGGTGTCTCCTGGCATTGACTCAACAACGACGACGGGGTTAACGAG |
| Length | 2549 |