CopciAB_165422
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_165422 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | STE7 | Synonyms | 165422 |
| Uniprot id | Functional description | Belongs to the protein kinase superfamily | |
| Location | scaffold_10:2025022..2027105 | Strand | - |
| Gene length (nt) | 2084 | Transcript length (nt) | 1827 |
| CDS length (nt) | 1308 | Protein length (aa) | 435 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7263982 | 65.7 | 4.703E-164 | 517 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | STE7 |
|---|---|
| Protein id | CopciAB_165422.T0 |
| Description | Belongs to the protein kinase superfamily |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd06620 | PKc_Byr1_like | - | 96 | 432 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 103 | 418 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR017441 | Protein kinase, ATP binding site |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K11226 |
EggNOG
| COG category | Description |
|---|---|
| T | Belongs to the protein kinase superfamily |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
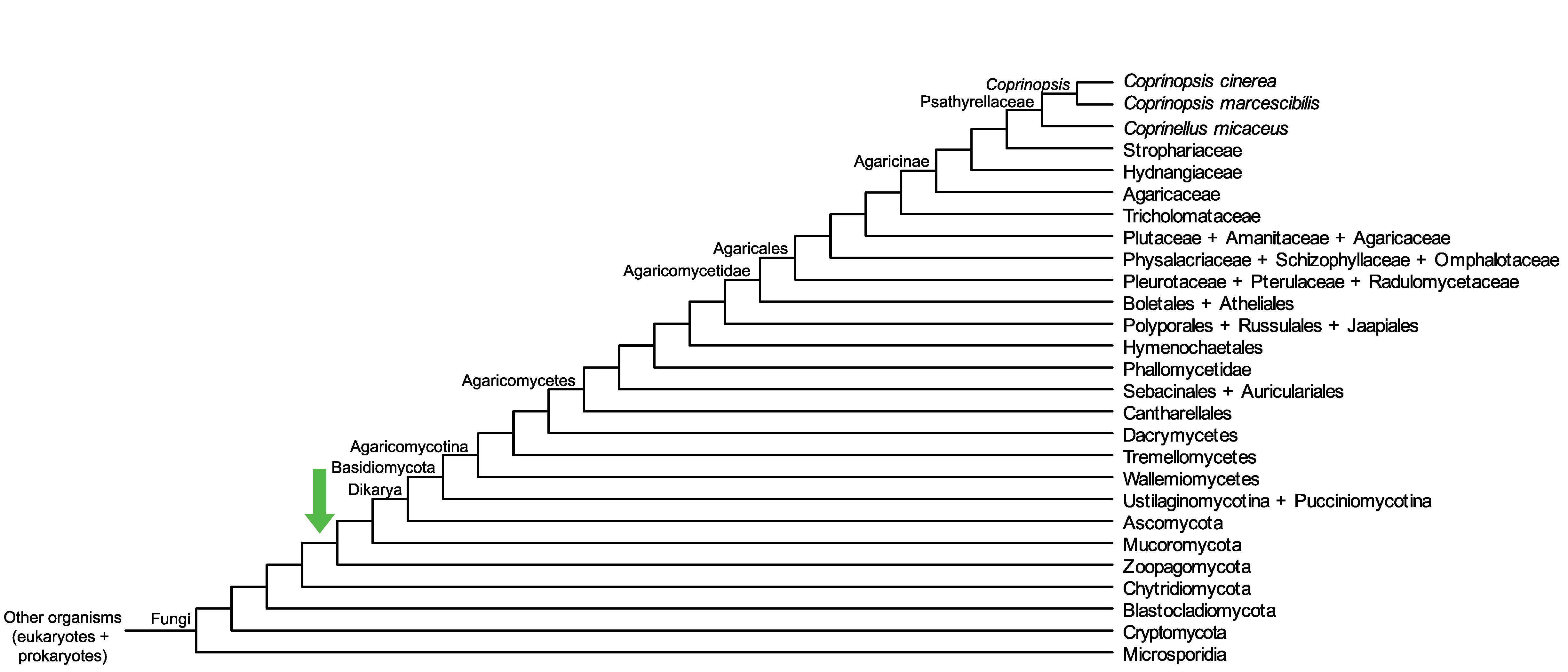
Conservation of CopciAB_165422 across fungi.
Arrow shows the origin of gene family containing CopciAB_165422.
Protein
| Sequence id | CopciAB_165422.T0 |
|---|---|
| Sequence |
>CopciAB_165422.T0 MTDLFVQASGPLRTKRNFKGLALSAEPPSPPPPGGKKRPPPLGAAMALGGGPAPLGGLPLLDDLPGESPATGRHS AMQATLSDKLAKLELKKRIKDLKNDDLKKLSDLGQGNGGSVEKVEHLPTGTIMAKKSVLIDAKSSVRKQILRELD IMHECQSDYIISCFGSFLAEPNICICMEFMDKGSFDGIYKKLGPLQVEVVGMVALSVLEGLTYLYDVHRIMHRDI KPSNILFNSQGQIKLCDFGVSGEVINSIANTFVGTSVYMSPERIQSHGDGYSVKSDVWSLGMSLVELALGAFPFT DPPDDDDDDLSDLEGDSSSPSPSDAQHQRTPTHRDSFLLSKKTAKRASKRRSKLTYNPDGQLSTMSIFELIHQIV QEPSPRLPSDKFPADAVDLVDICLLKNPEERQTPKALLNHPWIVAIRESDFDIQAFAATV |
| Length | 435 |
Coding
| Sequence id | CopciAB_165422.T0 |
|---|---|
| Sequence |
>CopciAB_165422.T0 ATGACAGACCTTTTCGTGCAAGCCTCAGGCCCTCTACGGACGAAACGAAACTTCAAAGGCCTCGCTCTGAGCGCG GAACCTCCTTCACCACCTCCTCCAGGCGGAAAGAAGCGACCACCGCCTCTCGGTGCGGCTATGGCACTCGGAGGT GGCCCAGCACCACTGGGCGGACTCCCACTTTTGGACGACCTACCAGGTGAAAGCCCCGCAACCGGCCGCCACAGC GCCATGCAAGCCACCCTCTCCGACAAGCTCGCCAAACTCGAACTCAAGAAACGGATAAAAGACCTCAAAAACGAC GATCTCAAGAAGCTCTCCGATCTGGGACAGGGTAATGGCGGAAGCGTAGAGAAAGTCGAACATCTCCCGACAGGA ACTATCATGGCTAAGAAGAGTGTTCTGATCGATGCCAAATCGTCCGTCAGGAAGCAGATCCTCCGCGAACTCGAC ATCATGCACGAGTGCCAATCCGACTACATCATCTCATGCTTTGGGTCGTTCCTGGCAGAACCCAATATCTGCATC TGTATGGAATTCATGGACAAAGGGTCGTTCGATGGGATCTACAAGAAACTGGGACCACTACAGGTCGAGGTTGTG GGGATGGTCGCGCTTTCCGTCTTGGAAGGCCTGACATATCTGTACGATGTTCATCGCATCATGCATCGCGACATC AAACCCTCAAACATCCTCTTCAACTCCCAAGGACAAATCAAACTATGCGACTTTGGCGTGTCCGGCGAAGTCATC AACTCCATCGCAAACACCTTCGTAGGGACATCCGTTTACATGAGCCCCGAACGCATCCAAAGCCACGGCGACGGC TACTCGGTCAAATCCGACGTCTGGTCCCTCGGCATGTCCCTCGTCGAACTCGCATTGGGCGCATTCCCCTTCACA GACCCACCAGACGACGACGACGACGACCTCTCCGACCTCGAAGGCGACTCCTCGAGTCCAAGTCCATCGGACGCC CAACACCAACGCACACCCACACACCGCGACTCGTTCCTCCTCAGCAAAAAGACCGCCAAACGCGCAAGCAAGCGC AGAAGCAAACTCACCTACAACCCAGATGGGCAGTTGTCCACCATGAGCATCTTTGAACTGATCCATCAAATCGTT CAGGAGCCCTCACCTCGACTCCCGAGCGACAAATTCCCAGCTGATGCCGTGGACCTGGTGGATATCTGTTTGTTG AAGAACCCAGAGGAGAGGCAAACGCCGAAAGCGCTATTGAACCATCCATGGATTGTCGCCATTCGAGAGTCAGAC TTTGACATTCAAGCCTTTGCCGCCACCGTTTAA |
| Length | 1308 |
Transcript
| Sequence id | CopciAB_165422.T0 |
|---|---|
| Sequence |
>CopciAB_165422.T0 GGTGCCCACTTCTTCTTCGTCACAACCTTGTTCTGAACCTAAATAACTTTTTCTAGCTTTATATCTTTTCGATTT CCAGCCCAACGACAACCAGCGCCCCACACCTCAACCCCAAAACCACCGGCACATCCGCAACAACATGACAGACCT TTTCGTGCAAGCCTCAGGCCCTCTACGGACGAAACGAAACTTCAAAGGCCTCGCTCTGAGCGCGGAACCTCCTTC ACCACCTCCTCCAGGCGGAAAGAAGCGACCACCGCCTCTCGGTGCGGCTATGGCACTCGGAGGTGGCCCAGCACC ACTGGGCGGACTCCCACTTTTGGACGACCTACCAGGTGAAAGCCCCGCAACCGGCCGCCACAGCGCCATGCAAGC CACCCTCTCCGACAAGCTCGCCAAACTCGAACTCAAGAAACGGATAAAAGACCTCAAAAACGACGATCTCAAGAA GCTCTCCGATCTGGGACAGGGTAATGGCGGAAGCGTAGAGAAAGTCGAACATCTCCCGACAGGAACTATCATGGC TAAGAAGAGTGTTCTGATCGATGCCAAATCGTCCGTCAGGAAGCAGATCCTCCGCGAACTCGACATCATGCACGA GTGCCAATCCGACTACATCATCTCATGCTTTGGGTCGTTCCTGGCAGAACCCAATATCTGCATCTGTATGGAATT CATGGACAAAGGGTCGTTCGATGGGATCTACAAGAAACTGGGACCACTACAGGTCGAGGTTGTGGGGATGGTCGC GCTTTCCGTCTTGGAAGGCCTGACATATCTGTACGATGTTCATCGCATCATGCATCGCGACATCAAACCCTCAAA CATCCTCTTCAACTCCCAAGGACAAATCAAACTATGCGACTTTGGCGTGTCCGGCGAAGTCATCAACTCCATCGC AAACACCTTCGTAGGGACATCCGTTTACATGAGCCCCGAACGCATCCAAAGCCACGGCGACGGCTACTCGGTCAA ATCCGACGTCTGGTCCCTCGGCATGTCCCTCGTCGAACTCGCATTGGGCGCATTCCCCTTCACAGACCCACCAGA CGACGACGACGACGACCTCTCCGACCTCGAAGGCGACTCCTCGAGTCCAAGTCCATCGGACGCCCAACACCAACG CACACCCACACACCGCGACTCGTTCCTCCTCAGCAAAAAGACCGCCAAACGCGCAAGCAAGCGCAGAAGCAAACT CACCTACAACCCAGATGGGCAGTTGTCCACCATGAGCATCTTTGAACTGATCCATCAAATCGTTCAGGAGCCCTC ACCTCGACTCCCGAGCGACAAATTCCCAGCTGATGCCGTGGACCTGGTGGATATCTGTTTGTTGAAGAACCCAGA GGAGAGGCAAACGCCGAAAGCGCTATTGAACCATCCATGGATTGTCGCCATTCGAGAGTCAGACTTTGACATTCA AGCCTTTGCCGCCACCGTTTAACCGCCGTTCTGCCGCACCTCACCGTCACCATCGCATCTGGACAATTGCATCTA TTGGGCTCAGGCTGGAACCCTTGTATCGATTGGGCTTCAGGCTGGAAACCAACCTAGCGCTCTGTACTATATGAA TAACTCTCTGTTCGGATTCGTGAATAATTCTTTGATCGTCTTCCCCAACCAATGCACCTTCTCATTATTGAACCT TCGTATCTATGCCCAAGCTCCATTCACCGACTGTTTGTTTTCTACCCACAACCGATTGCACGGTTGAACAATTAG CTCTGACACTGTTTCAGTTTTTCCTACCTCATGATCATCACTATACTTACCACCCCTGTACTGCTTACTGTGTAC TTCTATATAAGCCCCACATCGTTCTGT |
| Length | 1827 |
Gene
| Sequence id | CopciAB_165422.T0 |
|---|---|
| Sequence |
>CopciAB_165422.T0 GGTGCCCACTTCTTCTTCGTCACAACCTTGTTCTGAACCTAAATAACTTTTTCTAGCTTTATATCTTTTCGATTT CCAGCCCAACGACAACCAGCGCCCCACACCTCAACCCCAAAACCACCGGCACATCCGCAACAACATGACAGACCT TTTCGTGCAAGCCTCAGGCCCTCTACGGACGAAACGAAACTTCAAAGGCCTCGCTCTGAGCGCGGAACCTCCTTC ACCACCTCCTCCAGGCGGAAAGAAGCGACCACCGCCTCTCGGTGCGGCTATGGCACTCGGAGGTGGCCCAGCACC ACTGGGCGGACTCCCACTTTTGGACGACCTACCAGGTGAAAGCCCCGCAACCGGCCGCCACAGCGCCATGCAAGC CACCCTCTCCGACAAGCTCGCCAAACTCGAACTCAAGAAACGGATAAAAGACCTCAAAAACGACGATCTCAAGAA GCTCTCCGATCTGGGACAGGGTAATGGCGGAAGCGTAGAGAAAGTCGAACATCTCCCGACAGGAACTATCATGGC TAAGAAGGTGCGGGGTATCATTTAAACAACGATTTGTCGTTTTACGTCCTTGTATACGAGAAAGGCGCTGACGCT TGACATGTACACGTACAGAGTGTTCTGATCGATGCCAAATCGTCCGTCAGGAAGCAGATCCTCCGCGAACTCGAC ATCATGCACGAGTGCCAATCCGACTACATCATCTCATGCTTTGGGTCGTTCCTGGCAGAACCCAATATCTGCATC TGTATGGAATTCATGGACAAAGGGTCGTTCGATGGGATCTACAAGAAACTGGGACCACTACAGGTCGAGGTTGTG GGGATGGTCGCGCTTTCCGTCTTGGAAGGCCTGACATATCTGTACGATGTTCATCGCATCATGCATCGCGGTACG TCGCCATCTATGCCACTACTTATACCAGCGCTGACGGATATTATCTAGACATCAAACCCTCAAACATCCTCTTCA ACTCCCAAGGACAAATCAAACTATGCGACTTTGGCGTGTCCGGCGAAGTCATCAACTCCATCGCAAACACCTTCG TAGGGACATCCGTTTACATGAGCGTACGTTCCCATCCATCCATTCTCCACCCACTCCCAATTCCCTAACCTCTCG ACTCGTTAAGCCCGAACGCATCCAAAGCCACGGCGACGGCTACTCGGTCAAATCCGACGTCTGGTCCCTCGGCAT GTCCCTCGTCGAACTCGCATTGGGCGCATTCCCCTTCACAGACCCACCAGACGACGACGACGACGACCTCTCCGA CCTCGAAGGCGACTCCTCGAGTCCAAGTCCATCGGACGCCCAACACCAACGCACACCCACACACCGCGACTCGTT CCTCCTCAGCAAAAAGACCGCCAAACGCGCAAGCAAGCGCAGAAGCAAACTCACCTACAACCCAGATGGGCAGTT GTCCACCATGAGCATCTTTGAACTGATCCATCAAATCGTTCAGGAGCCCTCACCTCGACTCCCGAGCGACAAATT CCCAGCTGATGCCGTGGACCTGGTGGATATCTGTTTGTTGAAGAACCCAGAGGAGAGGCAAACGCCGAAAGCGCT ATTGGTGAGGATATATCTCGACTCCGAAGCTCGTGCTGATTTGAATCTATCGCCTCGTAGAACCATCCATGGATT GTCGCCATTCGAGAGTCAGACTTTGACATTCAAGCCTTTGCCGCCACCGTTTAACCGCCGTTCTGCCGCACCTCA CCGTCACCATCGCATCTGGACAATTGCATCTATTGGGCTCAGGCTGGAACCCTTGTATCGATTGGGCTTCAGGCT GGAAACCAACCTAGCGCTCTGTACTATATGAATAACTCTCTGTTCGGATTCGTGAATAATTCTTTGATCGTCTTC CCCAACCAATGCACCTTCTCATTATTGAACCTTCGTATCTATGCCCAAGCTCCATTCACCGACTGTTTGTTTTCT ACCCACAACCGATTGCACGGTTGAACAATTAGCTCTGACACTGTTTCAGTTTTTCCTACCTCATGATCATCACTA TACTTACCACCCCTGTACTGCTTACTGTGTACTTCTATATAAGCCCCACATCGTTCTGT |
| Length | 2084 |