CopciAB_232971
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_232971 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 232971 |
| Uniprot id | Functional description | Phosphotransferase enzyme family | |
| Location | scaffold_12:717807..721577 | Strand | - |
| Gene length (nt) | 3771 | Transcript length (nt) | 3450 |
| CDS length (nt) | 2475 | Protein length (aa) | 824 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_208685 | 58.9 | 5.712E-256 | 803 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_78050 | 59.4 | 5.952E-256 | 803 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB27484 | 56.8 | 5.047E-243 | 766 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_20182 | 55.1 | 4.257E-243 | 766 |
| Lentinula edodes NBRC 111202 | Lenedo1_1167597 | 56.3 | 1.023E-241 | 762 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1452418 | 54.7 | 1.064E-230 | 730 |
| Agrocybe aegerita | Agrae_CAA7268602 | 49 | 5.379E-230 | 728 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1098256 | 49.2 | 7.369E-223 | 707 |
| Schizophyllum commune H4-8 | Schco3_2635914 | 47.8 | 5.112E-221 | 702 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_117696 | 51.5 | 9.809E-220 | 698 |
| Flammulina velutipes | Flave_chr11AA00857 | 59.6 | 1.06E-218 | 695 |
| Lentinula edodes B17 | Lened_B_1_1_7159 | 54.4 | 1.904E-196 | 630 |
| Pleurotus ostreatus PC9 | PleosPC9_1_117137 | 54.1 | 2.811E-191 | 615 |
| Grifola frondosa | Grifr_OBZ77771 | 43.1 | 3.837E-175 | 568 |
| Auricularia subglabra | Aurde3_1_1274240 | 44 | 8.328E-121 | 409 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_232971.T0 |
| Description | Phosphotransferase enzyme family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF01636 | Phosphotransferase enzyme family | IPR002575 | 190 | 555 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR002575 | Aminoglycoside phosphotransferase |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | Phosphotransferase enzyme family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
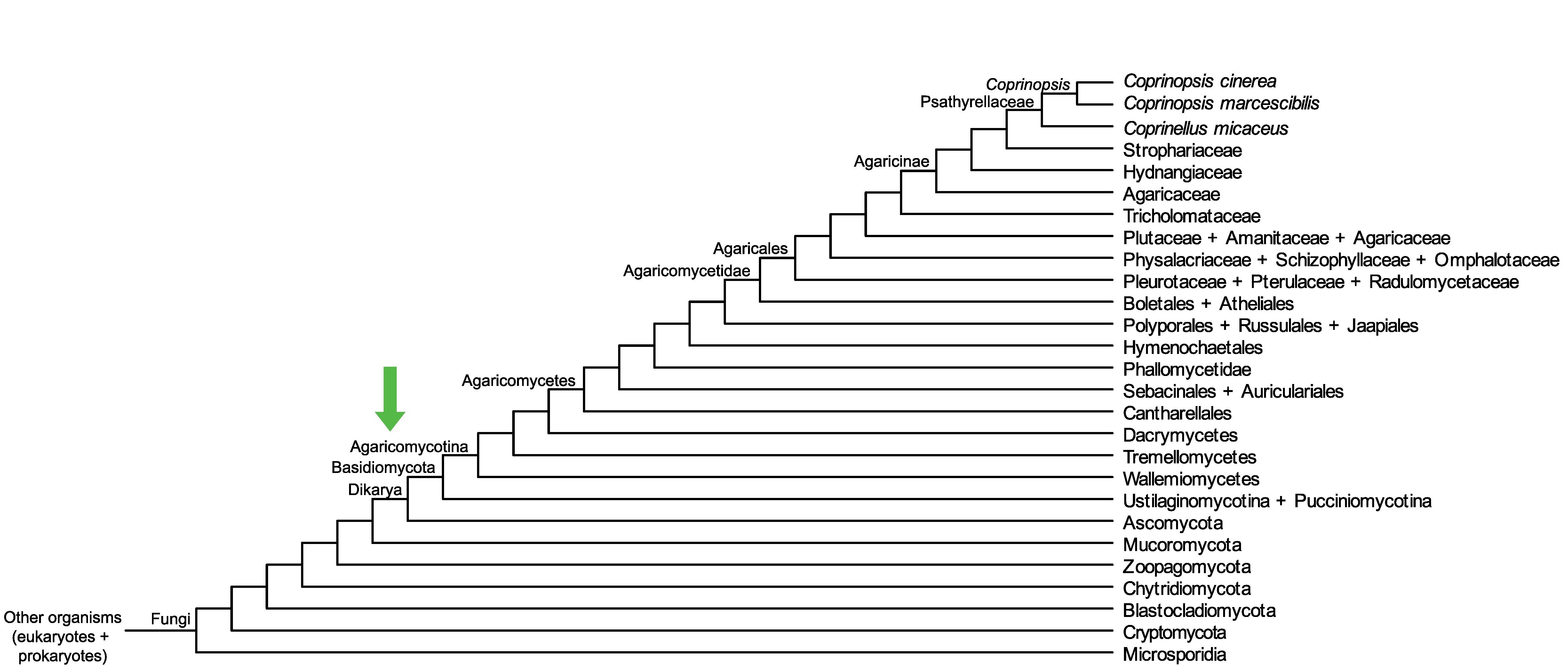
Conservation of CopciAB_232971 across fungi.
Arrow shows the origin of gene family containing CopciAB_232971.
Protein
| Sequence id | CopciAB_232971.T0 |
|---|---|
| Sequence |
>CopciAB_232971.T0 MGVLDMLHIDPCSNAVEHGLANIYKQAITGNNSLRVNFDYNAALARLSSLGSSSSGSNSVAVSSSLKGESSETSV DKNGHLDTTTATPASTDNDHPVMKKSGQKHLFNLVDASKTQDNPQPAKNPSPATVPPAPPAREPLEWDWDIEHED RKREKERDATLFPRETPFEVDRRLLKDVVREKMGVEVGRIKFLSAGTFHKAYLIILVNHEELVARVARRFMPRLK VESEVATISYLRQKTNVPVPKVYHYDSNPYNRLGGEFILMSKAPGIPLSRVYHGLAYNDLVKLVKNLARIVLPLF AHRFSDIGSLYFGPDPRANVASGTPTPKETQQHYSGFPFSPTLGMMTKSPSTQSVHTLSHQFHVGPIISWPFFGS NRGELVHPTEMNRGPWPTLKSYLESCVDREIKGVIRENEGRSAPHRLHLDPDEIRSSRHHKMRAVPGDESDDSDE YSDYEDADEDSEWPGSSIYSDFRRMQRTTFLVAHMNQREEAVRKEMGRWKSVMERLVDQVEKDGHREEFGLDCHD LSLENVFVDPEDNTKITCIIDWESTTTRPLWACAHVPACIQSSAFVSKLFREAVAELPSDPTFRVSLPQNGRTVD PAHLCREWLYYEAAGARLRMAHRCAEWDGWEEGLVDSILGPEEFESEWFKPGANEYDMEDLKAMIASPHGEEHQE LNGRSSSGRLSGSSKSSKASSSRSPAARNRSTTSSSKELMLSPKLTDVKNAVKNGVNGGGMGKLPPLLQEQQREK MLNQTGDICGGRGGELGLRLEAWLCEKEKGPKRWKGDGDEGGDGDDESEVQSPMTAGAHGAQTVHSVLTPRFER |
| Length | 824 |
Coding
| Sequence id | CopciAB_232971.T0 |
|---|---|
| Sequence |
>CopciAB_232971.T0 ATGGGTGTTCTCGACATGCTTCACATCGATCCTTGCTCGAATGCGGTTGAGCACGGTCTCGCAAACATCTACAAA CAAGCCATCACTGGAAACAATTCCCTGCGCGTCAATTTTGATTACAATGCCGCGTTGGCCAGGCTAAGCAGTCTT GGTTCCTCCAGTTCCGGATCGAACTCCGTTGCAGTTTCATCAAGTCTGAAAGGAGAATCGTCCGAAACGAGCGTT GATAAGAACGGTCATCTGGACACGACGACAGCGACCCCGGCCTCCACTGACAACGACCACCCTGTGATGAAGAAG AGTGGTCAAAAACACCTATTCAACCTCGTCGACGCTTCAAAAACTCAGGACAACCCACAACCTGCAAAAAATCCG TCACCAGCAACGGTACCACCAGCACCACCTGCCAGAGAACCCTTGGAGTGGGATTGGGATATCGAACATGAAGAC AGGAAACGAGAAAAGGAGAGAGATGCCACCTTGTTCCCCCGTGAAACCCCATTTGAGGTTGACCGGAGGCTGCTC AAGGATGTGGTCAGGGAGAAGATGGGTGTCGAGGTTGGGAGAATCAAATTCTTGAGTGCTGGAACTTTCCACAAG GCCTACCTCATCATCCTAGTGAACCACGAAGAACTCGTCGCCAGGGTCGCCAGGCGATTCATGCCCCGCCTCAAG GTCGAGTCTGAGGTGGCAACGATAAGCTATCTGCGCCAAAAGACCAACGTCCCCGTCCCCAAGGTCTACCACTAC GACTCGAACCCATATAACCGGTTAGGGGGCGAATTTATCCTCATGTCCAAGGCACCGGGCATTCCTCTATCACGC GTTTACCATGGCCTCGCCTACAACGATCTCGTCAAGCTTGTCAAGAACCTGGCCAGGATCGTGCTTCCCCTCTTC GCCCACCGCTTCTCTGATATCGGCTCCTTATACTTTGGACCCGACCCTCGAGCAAATGTAGCCTCCGGAACCCCG ACTCCCAAAGAGACCCAACAGCACTATTCCGGCTTCCCGTTTTCGCCTACTCTGGGCATGATGACCAAGTCCCCC TCCACCCAATCGGTTCACACCCTCTCCCACCAATTTCATGTGGGACCAATCATCTCGTGGCCCTTCTTTGGTTCT AACAGGGGAGAGCTCGTTCACCCAACGGAAATGAATCGTGGGCCATGGCCAACGCTCAAGTCCTACCTCGAGTCG TGTGTCGATCGAGAAATCAAGGGTGTTATTCGAGAGAATGAGGGTCGATCAGCGCCGCACCGGCTGCATCTCGAT CCTGACGAGATTCGTTCCTCGAGGCATCACAAGATGCGGGCTGTCCCCGGAGACGAGAGTGATGATTCGGACGAG TACAGTGATTACGAGGATGCCGACGAGGATTCGGAATGGCCGGGCAGTAGCATCTACTCTGATTTCCGGAGGATG CAGCGTACCACTTTCTTGGTTGCACATATGAACCAGCGAGAGGAGGCTGTCCGGAAAGAGATGGGGCGGTGGAAG AGCGTCATGGAGAGGCTAGTCGATCAAGTCGAGAAGGACGGCCACCGTGAAGAGTTTGGATTGGACTGCCACGAC CTTTCCCTCGAGAATGTCTTCGTCGACCCTGAAGACAACACCAAGATCACATGCATCATTGACTGGGAATCGACG ACGACGCGACCACTCTGGGCCTGCGCCCATGTTCCAGCTTGCATCCAGTCCAGCGCGTTTGTGTCCAAGTTGTTC CGGGAAGCTGTCGCCGAGCTTCCCTCCGACCCTACATTCCGAGTTTCGCTACCACAGAACGGGCGGACAGTCGAC CCTGCGCACTTGTGTCGGGAATGGCTGTATTACGAGGCGGCTGGAGCCCGGCTTCGCATGGCCCATCGATGTGCG GAGTGGGATGGATGGGAAGAGGGGTTGGTGGATAGTATCCTTGGACCTGAAGAGTTCGAATCCGAGTGGTTCAAG CCCGGAGCCAACGAGTACGATATGGAGGATCTAAAAGCTATGATCGCTTCTCCGCACGGCGAAGAGCACCAGGAA CTCAACGGGCGAAGTTCGAGTGGACGCTTGTCTGGGTCTTCGAAATCGTCGAAAGCGAGTTCGTCTCGCTCGCCG GCAGCGCGGAATCGGAGTACGACCAGCTCTTCGAAGGAGTTGATGCTTTCGCCCAAGCTCACCGACGTCAAGAAC GCTGTGAAGAATGGTGTGAATGGAGGGGGTATGGGCAAGTTGCCGCCATTATTGCAGGAGCAGCAGAGGGAGAAG ATGCTGAACCAGACGGGGGATATTTGTGGAGGGCGGGGCGGGGAGTTGGGGTTGAGGTTGGAGGCTTGGTTGTGT GAGAAGGAGAAGGGGCCGAAGAGGTGGAAGGGTGACGGCGATGAGGGTGGGGATGGGGATGATGAGAGTGAGGTG CAGTCGCCGATGACTGCTGGTGCTCATGGTGCCCAGACGGTTCACTCGGTGCTGACCCCGAGGTTTGAGCGATAG |
| Length | 2475 |
Transcript
| Sequence id | CopciAB_232971.T0 |
|---|---|
| Sequence |
>CopciAB_232971.T0 GGTTTCTCTTCCTTCCCTTCCCCGCAACGAACACCAGCAAAAAGTCTACAACTCTTCTTTCGTATGGTGCATGGG TGTTCTCGACATGCTTCACATCGATCCTTGCTCGAATGCGGTTGAGCACGGTCTCGCAAACATCTACAAACAAGC CATCACTGGAAACAATTCCCTGCGCGTCAATTTTGATTACAATGCCGCGTTGGCCAGGCTAAGCAGTCTTGGTTC CTCCAGTTCCGGATCGAACTCCGTTGCAGTTTCATCAAGTCTGAAAGGAGAATCGTCCGAAACGAGCGTTGATAA GAACGGTCATCTGGACACGACGACAGCGACCCCGGCCTCCACTGACAACGACCACCCTGTGATGAAGAAGAGTGG TCAAAAACACCTATTCAACCTCGTCGACGCTTCAAAAACTCAGGACAACCCACAACCTGCAAAAAATCCGTCACC AGCAACGGTACCACCAGCACCACCTGCCAGAGAACCCTTGGAGTGGGATTGGGATATCGAACATGAAGACAGGAA ACGAGAAAAGGAGAGAGATGCCACCTTGTTCCCCCGTGAAACCCCATTTGAGGTTGACCGGAGGCTGCTCAAGGA TGTGGTCAGGGAGAAGATGGGTGTCGAGGTTGGGAGAATCAAATTCTTGAGTGCTGGAACTTTCCACAAGGCCTA CCTCATCATCCTAGTGAACCACGAAGAACTCGTCGCCAGGGTCGCCAGGCGATTCATGCCCCGCCTCAAGGTCGA GTCTGAGGTGGCAACGATAAGCTATCTGCGCCAAAAGACCAACGTCCCCGTCCCCAAGGTCTACCACTACGACTC GAACCCATATAACCGGTTAGGGGGCGAATTTATCCTCATGTCCAAGGCACCGGGCATTCCTCTATCACGCGTTTA CCATGGCCTCGCCTACAACGATCTCGTCAAGCTTGTCAAGAACCTGGCCAGGATCGTGCTTCCCCTCTTCGCCCA CCGCTTCTCTGATATCGGCTCCTTATACTTTGGACCCGACCCTCGAGCAAATGTAGCCTCCGGAACCCCGACTCC CAAAGAGACCCAACAGCACTATTCCGGCTTCCCGTTTTCGCCTACTCTGGGCATGATGACCAAGTCCCCCTCCAC CCAATCGGTTCACACCCTCTCCCACCAATTTCATGTGGGACCAATCATCTCGTGGCCCTTCTTTGGTTCTAACAG GGGAGAGCTCGTTCACCCAACGGAAATGAATCGTGGGCCATGGCCAACGCTCAAGTCCTACCTCGAGTCGTGTGT CGATCGAGAAATCAAGGGTGTTATTCGAGAGAATGAGGGTCGATCAGCGCCGCACCGGCTGCATCTCGATCCTGA CGAGATTCGTTCCTCGAGGCATCACAAGATGCGGGCTGTCCCCGGAGACGAGAGTGATGATTCGGACGAGTACAG TGATTACGAGGATGCCGACGAGGATTCGGAATGGCCGGGCAGTAGCATCTACTCTGATTTCCGGAGGATGCAGCG TACCACTTTCTTGGTTGCACATATGAACCAGCGAGAGGAGGCTGTCCGGAAAGAGATGGGGCGGTGGAAGAGCGT CATGGAGAGGCTAGTCGATCAAGTCGAGAAGGACGGCCACCGTGAAGAGTTTGGATTGGACTGCCACGACCTTTC CCTCGAGAATGTCTTCGTCGACCCTGAAGACAACACCAAGATCACATGCATCATTGACTGGGAATCGACGACGAC GCGACCACTCTGGGCCTGCGCCCATGTTCCAGCTTGCATCCAGTCCAGCGCGTTTGTGTCCAAGTTGTTCCGGGA AGCTGTCGCCGAGCTTCCCTCCGACCCTACATTCCGAGTTTCGCTACCACAGAACGGGCGGACAGTCGACCCTGC GCACTTGTGTCGGGAATGGCTGTATTACGAGGCGGCTGGAGCCCGGCTTCGCATGGCCCATCGATGTGCGGAGTG GGATGGATGGGAAGAGGGGTTGGTGGATAGTATCCTTGGACCTGAAGAGTTCGAATCCGAGTGGTTCAAGCCCGG AGCCAACGAGTACGATATGGAGGATCTAAAAGCTATGATCGCTTCTCCGCACGGCGAAGAGCACCAGGAACTCAA CGGGCGAAGTTCGAGTGGACGCTTGTCTGGGTCTTCGAAATCGTCGAAAGCGAGTTCGTCTCGCTCGCCGGCAGC GCGGAATCGGAGTACGACCAGCTCTTCGAAGGAGTTGATGCTTTCGCCCAAGCTCACCGACGTCAAGAACGCTGT GAAGAATGGTGTGAATGGAGGGGGTATGGGCAAGTTGCCGCCATTATTGCAGGAGCAGCAGAGGGAGAAGATGCT GAACCAGACGGGGGATATTTGTGGAGGGCGGGGCGGGGAGTTGGGGTTGAGGTTGGAGGCTTGGTTGTGTGAGAA GGAGAAGGGGCCGAAGAGGTGGAAGGGTGACGGCGATGAGGGTGGGGATGGGGATGATGAGAGTGAGGTGCAGTC GCCGATGACTGCTGGTGCTCATGGTGCCCAGACGGTTCACTCGGTGCTGACCCCGAGGTTTGAGCGATAGGAAGG GGGGCGTGTGGATGAACGACTCGGCCCTGTTCTGGTTCGGGATCGAGACCCGGCCTGACTTCTACCTCTTTGCCT TCCATCTCATATGCGAAAGATAGAAGGAAGGAACGGACGAACGAACGACCGAATGAACGCTTTCATTGGCGCTGT TCTAAGTTCTGGTTGGTTGACGAACTGTTATTTTATTTATCCTCATCATCATTGCTTGCATTGTTTCAACTCTCG TTTTTCGCATTGGCATTCAACTGTTCATGATTGTTAACGTTCACTTCACGTATTTTGGTTGGGGGTTACCTTTCG TCCATTCTTTCCTCTTCAGTCTTCTTCGTCCATCGTCTTTCGGCATTGGCATCGTCATCTTCCGTCGTTGTCTTC GCATCATCGTCGGTTATTATTTCTTGGTTGTTGAATCATTTAAACTTGTCCCTGGTTTTTGTCTCATGTCGGCTA CACGTTCAACATTCGCTGTGACACTGGGTTTCTTTTTTCCCCTCTCGTTCGCCGGGTAGATTAGCCTCGCCTTAT ATTTTTCACGCGAAGGTTATCTACGTAGCGTTTTTGCTTGCGTTGGCCATATAGGCACACATACATATACATTTT TACGAGTATTTTTTGTTGCTTTTGACGAGTGTCTTTTGGCGGTGGTTGGGTGGAGTGGTTTGTGGAGAGGAGGTG GGGGTGGAGGTGGACGGAGGAGGCAGATCTGAGGATACAGTCCAACTGGCGGGAGCAAGGAAACTAAGAAGTCCA TATCAAATCAAATCAAGTCTCAATCTCAAGTTTTTTCATCTACCACTACTATCTCTACTACTCATTTTATTGCTC TAGTGTTTTTTTCGGCGTATACGTACGCCTTGTAGCTCTTTTAGCATCGCATTATAAATCAATTTTATCATGGTT |
| Length | 3450 |
Gene
| Sequence id | CopciAB_232971.T0 |
|---|---|
| Sequence |
>CopciAB_232971.T0 GGTTTCTCTTCCTTCCCTTCCCCGCAACGAACACCAGCAAAAAGTCTACAACTCTTCTTTCGTATGGTGCATGGG TGTTCTCGACATGCTTCACATCGATCCTTGCTCGAATGCGGTTGAGCACGGTCTCGCAAACATCTACAAACAAGC CATCACTGGAAACAATTCCCTGCGCGTCAATTTTGATTACAAGTGCGTATTATTTCTCTTCATCCTTCATCGTCC AGAAATCTTTTTGTCTTGCTCGGAACCACACAGTCCTGACATTTTCTGCAGTGCCGCGTTGGCCAGGCTAAGCAG TCTTGGTTCCTCCAGTTCCGGATCGAACTCCGTTGCAGTTTCATCAAGTCTGAAAGGAGAATCGTCCGAAACGAG CGTTGATAAGAACGGTCATCTGGACACGACGACAGCGACCCCGGCCTCCACTGACAACGACCACCCTGTGATGAA GAAGAGTGGTCAAAAACACCTATTCAACCTCGTCGACGCTTCAAAAACTCAGGACAACCCACAACCTGCAAAAAA TCCGTCACCAGCAACGGTACCACCAGCACCACCTGCCAGAGAACCCTTGGAGTGGGATTGGGATATCGAACATGA AGACAGGAAACGAGAAAAGGAGAGAGATGCCACCTTGTTCCCCCGTGAAACCCCATTTGAGGTTGACCGGAGGCT GCTCAAGGATGTGGTCAGGGAGAAGATGGGTGTCGAGGTTGGGAGAATCAAATTCTTGAGTGCTGGTGCGTCGCT TTGCCTTTACCTTTGATCTTTTAAGCTCATACCGCGCCCAGGAACTTTCCACAAGGTTGACACGTCGACTTTTTC CTGATTTTCTGCCAGCTGACACCGTTACAGGCCTACCTCATCATCCTAGTGAACCACGAAGAACTCGTCGCCAGG GTCGCCAGGCGATTCATGCCCCGCCTCAAGGTCGAGTCTGAGGTGGCAACGATAAGCTATCTGCGCCAAAAGACC AACGTCCCCGTCCCCAAGGTCTACCACTACGACTCGAACCCATATAACCGGTTAGGGGGCGAATTTATCCTCATG TCCAAGGTACGCCAATTGTCCCTACCGCGTACCTTCTTTGGCTCAGTCACGTGCTGTGAAGGCACCGGGCATTCC TCTATCACGCGTTTACCATGGCCTCGCCTACAACGATCTCGTCAAGCTTGTCAAGAACCTGGCCAGGATCGTGCT TCCCCTCTTCGCCCACCGCTTCTCTGATATCGGCTCCTTATACTTTGGACCCGACCCTCGAGCAAATGTAGCCTC CGGAACCCCGACTCCCAAAGAGACCCAACAGCACTATTCCGGCTTCCCGTTTTCGCCTACTCTGGGCATGATGAC CAAGTCCCCCTCCACCCAATCGGTTCACACCCTCTCCCACCAATTTCATGTGGGACCAATCATCTCGTGGCCCTT CTTTGGTTCTAACAGGGGAGAGCTCGTTCACCCAACGGAAATGAATCGTGGGCCATGGCCAACGCTCAAGTCCTA CCTCGAGTCGTGTGTCGATCGAGAAATCAAGGGTGTTATTCGAGAGAATGAGGGTCGATCAGCGCCGCACCGGCT GCATCTCGATCCTGACGAGATTCGTTCCTCGAGGCATCACAAGATGCGGGCTGTCCCCGGAGACGAGAGTGATGA TTCGGACGAGTACAGTGATTACGAGGATGCCGACGAGGATTCGGAATGGCCGGGCAGTAGCATCTACTCTGATTT CCGGAGGATGCAGCGTACCACTTTCTTGGTTGCACATATGAACCAGCGAGAGGAGGCTGTCCGGAAAGAGATGGG GCGGTGGAAGAGCGTCATGGAGAGGCTAGTCGATCAAGTCGAGAAGGACGGCCACCGTGAAGAGTTTGGATTGGA CTGCCACGACCTTTCCCTCGAGAATGTCTTCGTCGACCCTGAAGACAACACCAAGATCGTAAGTAACCGACCCCT ATTCATATGCGATGGTTAACCAATCTAATCACTCTTTTGGTTCTTTTTCGTTGCCACGTTGTAGACATGCATCAT TGACTGGGAATCGACGACGACGCGACCACTCTGGGCCTGCGCCCATGTTCCAGCTTGCATCCAGTCCAGCGCGTT TGTGTCCAAGTTGTTCCGGGAAGCTGTCGCCGAGCTTCCCTCCGACCCTACATTCCGAGTTTCGCTACCACAGAA CGGGCGGACAGTCGACCCTGCGCACTTGTGTCGGGAATGGCTGTATTACGAGGCGGCTGGAGCCCGGCTTCGCAT GGCCCATCGATGTGCGGAGTGGGATGGATGGGAAGAGGGGTTGGTGGATAGTATCCTTGGACCTGAAGAGTTCGA ATCCGAGTGGTTCAAGCCCGGAGCCAACGAGTACGATATGGAGGATCTAAAAGCTATGATCGCTTCTCCGCACGG CGAAGAGCACCAGGAACTCAACGGGCGAAGTTCGAGTGGACGCTTGTCTGGGTCTTCGAAATCGTCGAAAGCGAG TTCGTCTCGCTCGCCGGCAGCGCGGAATCGGAGTACGACCAGCTCTTCGAAGGAGTTGATGCTTTCGCCCAAGCT CACCGACGTCAAGAACGCTGTGAAGAATGGTGTGAATGGAGGGGGTATGGGCAAGTTGCCGCCATTATTGCAGGA GCAGCAGAGGGAGAAGATGCTGAACCAGACGGGGGATATTTGTGGAGGGCGGGGCGGGGAGTTGGGGTTGAGGTT GGAGGCTTGGTTGTGTGAGAAGGAGAAGGGGCCGAAGAGGTGGAAGGGTGACGGCGATGAGGGTGGGGATGGGGA TGATGAGAGTGAGGTGCAGTCGCCGATGACTGCTGGTGCTCATGGTGCCCAGACGGTTCACTCGGTGCTGACCCC GAGGTTTGAGCGATAGGAAGGGGGGCGTGTGGATGAACGACTCGGCCCTGTTCTGGTTCGGGATCGAGACCCGGC CTGACTTCTACCTCTTTGCCTTCCATCTCATATGCGAAAGATAGAAGGAAGGAACGGACGAACGAACGACCGAAT GAACGCTTTCATTGGCGCTGTTCTAAGTTCTGGTTGGTTGACGAACTGTTATTTTATTTATCCTCATCATCATTG CTTGCATTGTTTCAACTCTCGTTTTTCGCATTGGCATTCAACTGTTCATGATTGTTAACGTTCACTTCACGTATT TTGGTTGGGGGTTACCTTTCGTCCATTCTTTCCTCTTCAGTCTTCTTCGTCCATCGTCTTTCGGCATTGGCATCG TCATCTTCCGTCGTTGTCTTCGCATCATCGTCGGTTATTATTTCTTGGTTGTTGAATCATTTAAACTTGTCCCTG GTTTTTGTCTCATGTCGGCTACACGTTCAACATTCGCTGTGACACTGGGTTTCTTTTTTCCCCTCTCGTTCGCCG GGTAGATTAGCCTCGCCTTATATTTTTCACGCGAAGGTTATCTACGTAGCGTTTTTGCTTGCGTTGGCCATATAG GCACACATACATATACATTTTTACGAGTATTTTTTGTTGCTTTTGACGAGTGTCTTTTGGCGGTGGTTGGGTGGA GTGGTTTGTGGAGAGGAGGTGGGGGTGGAGGTGGACGGAGGAGGCAGATCTGAGGATACAGTCCAACTGGCGGGA GCAAGGAAACTAAGAAGTCCATATCAAATCAAATCAAGTCTCAATCTCAAGTTTTTTCATCTACCACTACTATCT CTACTACTCATTTTATTGCTCTAGTGTTTTTTTCGGCGTATACGTACGCCTTGTAGCTCTTTTAGCATCGCATTA TAAATCAATTTTATCATGGTT |
| Length | 3771 |