CopciAB_261683
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_261683 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 261683 |
| Uniprot id | Functional description | Tkl lisk lisk-dd1 protein kinase | |
| Location | scaffold_10:1349614..1351726 | Strand | + |
| Gene length (nt) | 2113 | Transcript length (nt) | 1908 |
| CDS length (nt) | 1824 | Protein length (aa) | 607 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262574 | 68.5 | 4.081E-270 | 833 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB19657 | 66.2 | 7.092E-265 | 818 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1475170 | 66.1 | 2.076E-259 | 802 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_13254 | 63.2 | 5.262E-248 | 769 |
| Lentinula edodes NBRC 111202 | Lenedo1_1162820 | 62.8 | 1.395E-245 | 762 |
| Flammulina velutipes | Flave_chr04AA00387 | 65.4 | 2.854E-245 | 761 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_210461 | 63.1 | 1.451E-241 | 750 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_120216 | 61.8 | 2.323E-238 | 741 |
| Grifola frondosa | Grifr_OBZ74830 | 62.4 | 3.256E-234 | 729 |
| Lentinula edodes B17 | Lened_B_1_1_1918 | 58.3 | 1.123E-226 | 707 |
| Auricularia subglabra | Aurde3_1_1277550 | 58.6 | 6.243E-214 | 671 |
| Schizophyllum commune H4-8 | Schco3_2071878 | 68.7 | 2.062E-184 | 585 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1094603 | 78.2 | 9.634E-181 | 574 |
| Pleurotus ostreatus PC9 | PleosPC9_1_39251 | 78 | 9.966E-170 | 542 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_12916 | 77.5 | 2.541E-166 | 532 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_261683.T0 |
| Description | Tkl lisk lisk-dd1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd13999 | STKc_MAP3K-like | - | 21 | 270 |
| CDD | cd00029 | C1 | - | 544 | 581 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 19 | 268 |
| Pfam | PF00130 | Phorbol esters/diacylglycerol binding domain (C1 domain) | IPR002219 | 545 | 583 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR046349 | C1-like domain superfamily |
| IPR002219 | Protein kinase C-like, phorbol ester/diacylglycerol-binding domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
| IPR000719 | Protein kinase domain |
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| K05743 |
EggNOG
| COG category | Description |
|---|---|
| G | Tkl lisk lisk-dd1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
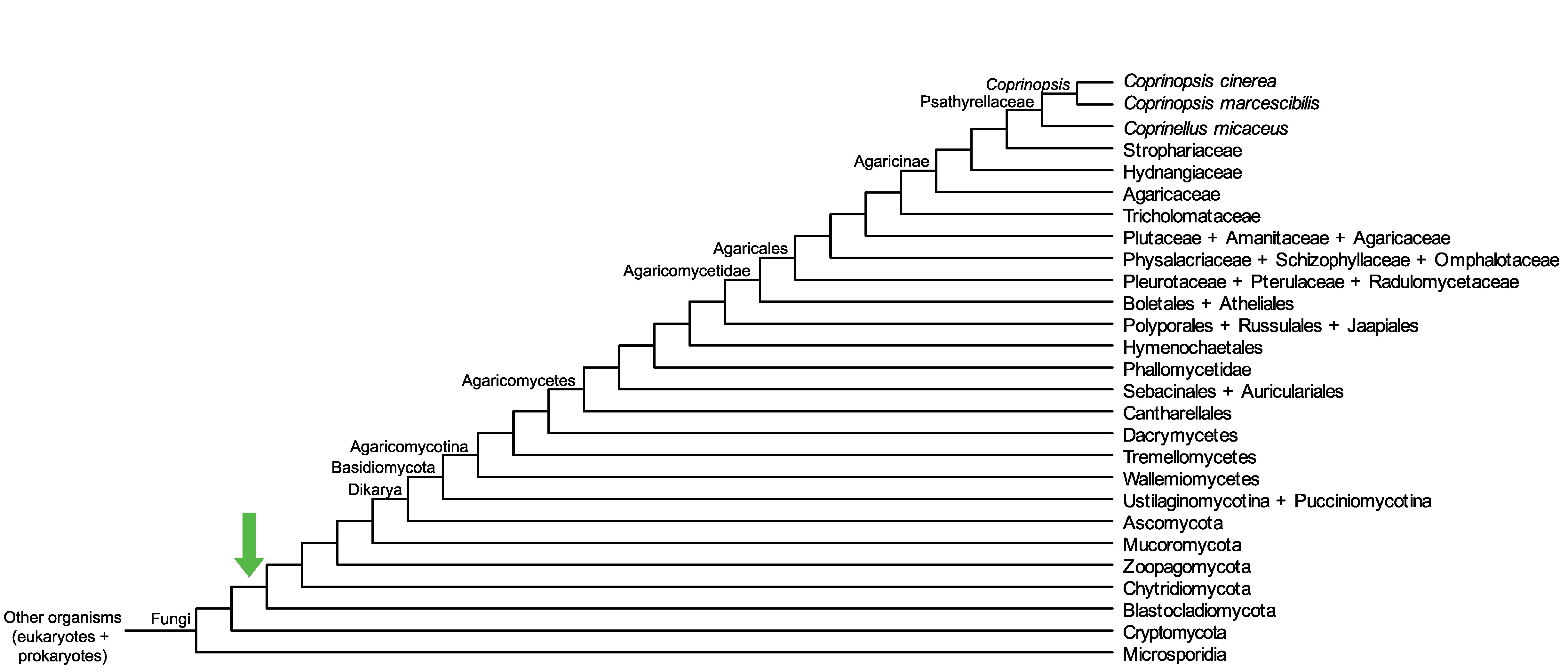
Conservation of CopciAB_261683 across fungi.
Arrow shows the origin of gene family containing CopciAB_261683.
Protein
| Sequence id | CopciAB_261683.T0 |
|---|---|
| Sequence |
>CopciAB_261683.T0 MHSDAFDVIPYEDIKGDWKKLGSGSFGNVYKGSYLGIDVAIKEVLPSNDYDVAKYFEREWRLMKECRHPNICLFI GLSRAPPPDNRIFIVSEFIEGGNVRLYIHDKNKPFPWRLRMSFATDVARALAYLHARKCIHRDLKGENLLVTSNG RIKVTDFGFARIAARNEEELRRLTFCGTDSYMSPEILLGDEFDLPTDVFSLGIIFCEIAARRLADDRHFKRHPPT FGIDPEEVRKLATPGCPEDFLKLCLDCLNTEPARRPGTREILDRLRLIEAEVLARPAEDGDVNVGSVRFISGARR PGAAPRIPSFGMGVGKDIRGGSGMSSASSASSDESDEELMEAVSRLTITKAGLEERSAWSTSSSVQPLLDNQHTP NGSTYSTSVIRQAAHSNTPSAPALSSILTIRPSPDPNRVPEHVLPATTSDISHASTASVDPLESPSIVSIATLDS YRTANTGESVISSTYATEGGSTIRSLHAPGHAVLPTVHRFTLLKPGTKPKKVSLGGGPGGIGGAGGTAGVVTQSS VETSVWNPLEIFFSSGLLISKCDLCAKRIGWKPFLECDDCGLRTHVKCGDLAPRDCGMRAVMNGHQHSTPVEMKK ISQSDSK |
| Length | 607 |
Coding
| Sequence id | CopciAB_261683.T0 |
|---|---|
| Sequence |
>CopciAB_261683.T0 ATGCATTCAGATGCCTTTGATGTTATTCCCTACGAGGACATCAAGGGGGATTGGAAGAAACTAGGTTCAGGCTCT TTTGGCAACGTCTACAAGGGCAGTTACCTGGGCATCGATGTGGCGATCAAAGAGGTTCTCCCATCCAACGACTAC GACGTTGCAAAATACTTTGAACGAGAATGGAGGCTCATGAAGGAATGCAGGCATCCCAACATTTGTCTTTTCATT GGCCTCAGCCGCGCTCCTCCACCCGATAACCGCATCTTCATCGTCTCAGAGTTCATCGAAGGAGGAAATGTCCGC CTCTATATCCACGACAAGAACAAACCCTTCCCCTGGCGCCTCAGAATGTCCTTTGCAACAGACGTCGCACGCGCA CTCGCTTACCTCCATGCCCGCAAGTGTATCCACCGCGATCTCAAGGGAGAGAATCTCCTTGTCACTTCCAATGGT CGCATCAAGGTCACCGACTTTGGTTTCGCCAGGATTGCAGCTAGGAACGAGGAGGAGCTGAGACGTCTCACCTTT TGTGGCACCGATTCCTACATGTCCCCTGAAATCCTCCTTGGCGACGAGTTTGATCTCCCTACCGATGTCTTCTCC CTTGGGATCATCTTTTGCGAGATTGCGGCGAGAAGGTTGGCGGACGATAGGCATTTCAAGCGTCACCCACCGACG TTTGGAATTGACCCCGAGGAGGTGAGAAAGCTTGCTACCCCTGGCTGTCCGGAGGACTTTTTGAAGCTATGTTTG GATTGCTTGAATACCGAGCCAGCGAGGAGGCCTGGGACGAGGGAAATATTGGACAGGTTGAGGTTAATCGAAGCG GAGGTACTTGCGAGGCCTGCCGAGGATGGTGATGTCAATGTGGGGAGCGTTAGGTTTATTTCGGGCGCCCGCAGG CCCGGGGCCGCACCTAGGATTCCGAGCTTTGGTATGGGAGTTGGGAAGGATATCAGAGGTGGCAGTGGTATGAGC AGTGCCAGTAGCGCCAGCTCCGATGAGAGTGACGAGGAGCTGATGGAGGCGGTTTCGAGGTTAACGATTACCAAG GCCGGATTGGAGGAAAGGTCGGCCTGGAGTACTTCATCTAGTGTACAACCTCTACTCGACAATCAACATACGCCT AATGGATCAACCTATAGCACCTCCGTTATCCGTCAAGCTGCACATTCTAATACGCCCTCCGCACCCGCTCTATCC TCAATCCTGACTATCAGACCTTCACCCGATCCTAATCGTGTCCCTGAACACGTCCTCCCGGCGACAACGTCCGAC ATATCTCACGCATCTACTGCGAGTGTGGACCCTCTGGAATCACCTTCGATCGTGTCCATCGCAACCCTCGACTCG TATCGCACAGCCAACACAGGTGAAAGCGTCATTTCAAGCACGTATGCAACTGAGGGTGGCTCCACCATTCGATCG CTGCATGCTCCTGGACATGCAGTCCTTCCGACTGTACATCGCTTCACATTGTTGAAGCCCGGTACAAAGCCGAAG AAGGTGTCGTTGGGTGGAGGACCGGGAGGTATAGGAGGGGCGGGAGGTACTGCTGGGGTGGTCACCCAGAGTTCT GTCGAAACCTCGGTCTGGAACCCGCTGGAGATATTCTTCTCGAGCGGGTTATTGATTTCGAAATGTGACCTTTGT GCGAAGAGGATTGGGTGGAAGCCGTTTTTGGAGTGCGATGATTGTGGGTTGAGGACCCACGTCAAGTGCGGTGAT CTGGCCCCGAGGGATTGTGGGATGCGAGCAGTTATGAATGGGCATCAACATTCGACGCCTGTTGAGATGAAGAAG ATTTCTCAGTCGGATTCGAAGTAG |
| Length | 1824 |
Transcript
| Sequence id | CopciAB_261683.T0 |
|---|---|
| Sequence |
>CopciAB_261683.T0 AGTCTCTTATTTCACTCCTTTCACCACTCTCACCTATACCATGCATTCAGATGCCTTTGATGTTATTCCCTACGA GGACATCAAGGGGGATTGGAAGAAACTAGGTTCAGGCTCTTTTGGCAACGTCTACAAGGGCAGTTACCTGGGCAT CGATGTGGCGATCAAAGAGGTTCTCCCATCCAACGACTACGACGTTGCAAAATACTTTGAACGAGAATGGAGGCT CATGAAGGAATGCAGGCATCCCAACATTTGTCTTTTCATTGGCCTCAGCCGCGCTCCTCCACCCGATAACCGCAT CTTCATCGTCTCAGAGTTCATCGAAGGAGGAAATGTCCGCCTCTATATCCACGACAAGAACAAACCCTTCCCCTG GCGCCTCAGAATGTCCTTTGCAACAGACGTCGCACGCGCACTCGCTTACCTCCATGCCCGCAAGTGTATCCACCG CGATCTCAAGGGAGAGAATCTCCTTGTCACTTCCAATGGTCGCATCAAGGTCACCGACTTTGGTTTCGCCAGGAT TGCAGCTAGGAACGAGGAGGAGCTGAGACGTCTCACCTTTTGTGGCACCGATTCCTACATGTCCCCTGAAATCCT CCTTGGCGACGAGTTTGATCTCCCTACCGATGTCTTCTCCCTTGGGATCATCTTTTGCGAGATTGCGGCGAGAAG GTTGGCGGACGATAGGCATTTCAAGCGTCACCCACCGACGTTTGGAATTGACCCCGAGGAGGTGAGAAAGCTTGC TACCCCTGGCTGTCCGGAGGACTTTTTGAAGCTATGTTTGGATTGCTTGAATACCGAGCCAGCGAGGAGGCCTGG GACGAGGGAAATATTGGACAGGTTGAGGTTAATCGAAGCGGAGGTACTTGCGAGGCCTGCCGAGGATGGTGATGT CAATGTGGGGAGCGTTAGGTTTATTTCGGGCGCCCGCAGGCCCGGGGCCGCACCTAGGATTCCGAGCTTTGGTAT GGGAGTTGGGAAGGATATCAGAGGTGGCAGTGGTATGAGCAGTGCCAGTAGCGCCAGCTCCGATGAGAGTGACGA GGAGCTGATGGAGGCGGTTTCGAGGTTAACGATTACCAAGGCCGGATTGGAGGAAAGGTCGGCCTGGAGTACTTC ATCTAGTGTACAACCTCTACTCGACAATCAACATACGCCTAATGGATCAACCTATAGCACCTCCGTTATCCGTCA AGCTGCACATTCTAATACGCCCTCCGCACCCGCTCTATCCTCAATCCTGACTATCAGACCTTCACCCGATCCTAA TCGTGTCCCTGAACACGTCCTCCCGGCGACAACGTCCGACATATCTCACGCATCTACTGCGAGTGTGGACCCTCT GGAATCACCTTCGATCGTGTCCATCGCAACCCTCGACTCGTATCGCACAGCCAACACAGGTGAAAGCGTCATTTC AAGCACGTATGCAACTGAGGGTGGCTCCACCATTCGATCGCTGCATGCTCCTGGACATGCAGTCCTTCCGACTGT ACATCGCTTCACATTGTTGAAGCCCGGTACAAAGCCGAAGAAGGTGTCGTTGGGTGGAGGACCGGGAGGTATAGG AGGGGCGGGAGGTACTGCTGGGGTGGTCACCCAGAGTTCTGTCGAAACCTCGGTCTGGAACCCGCTGGAGATATT CTTCTCGAGCGGGTTATTGATTTCGAAATGTGACCTTTGTGCGAAGAGGATTGGGTGGAAGCCGTTTTTGGAGTG CGATGATTGTGGGTTGAGGACCCACGTCAAGTGCGGTGATCTGGCCCCGAGGGATTGTGGGATGCGAGCAGTTAT GAATGGGCATCAACATTCGACGCCTGTTGAGATGAAGAAGATTTCTCAGTCGGATTCGAAGTAGTACATATAATG TAGTTGTAGATGCAATTGGACTAGTAATGTGTA |
| Length | 1908 |
Gene
| Sequence id | CopciAB_261683.T0 |
|---|---|
| Sequence |
>CopciAB_261683.T0 AGTCTCTTATTTCACTCCTTTCACCACTCTCACCTATACCATGCATTCAGATGCCTTTGATGTTATTCCCTACGA GGTCCTCTTATTTCTCAAACGATCTATACCACACACTCATATATCCACATAGGACATCAAGGGGGATTGGAAGAA ACTAGGTTCAGGCTCTTTTGGCAACGTCTACAAGGGTGCGTCTTCCCTGAATCTATTGCGAATTTATCTCCAATT GTCTATGCAGGCAGTTACCTGGGCATCGATGTGGCGATCAAAGAGGTTCTCCCATCCAACGACTACGACGTTGCA AAATACTTTGAACGAGAATGGAGGCTCATGAAGGAATGCAGGCATCCCAACATTTGTCTTTTCATTGGCCTCAGC CGCGCTCCTCCACCCGATAACCGCATCTTCATCGTCTCAGAGTTCATCGAAGGAGGAAATGTCCGCCTCTATATC CACGACAAGAACAAACCCTTCCCCTGGCGCCTCAGAATGTCCTTTGCAACAGACGTCGCACGCGCACTCGCTTAC CTCCATGCCCGCAAGTGTATCCACCGCGATCTCAAGGGAGAGAATCTCCTTGTCACTTCCAATGGTCGCATCAAG GTCACCGACTTTGGTTTCGCCAGGATTGCAGCTAGGAACGAGGAGGAGCTGAGACGTCTCACCTTTTGTGGCACC GATTCCTACATGTCCCCTGAAATCCTCCTTGGCGACGAGTTTGATCTCCCTACCGATGTCTTCTCCCTTGGGATC ATCTTTTGCGAGATTGCGGCGAGAAGGTTGGCGGACGATAGGCATTTCAAGCGTCACCCACCGACGTTTGGAATT GACCCCGAGGAGGTGAGAAAGCTTGCTACCCCTGGCTGTCCGGAGGACTTTTTGAAGCTATGTTTGGATTGCTTG AATACCGAGCCAGCGAGGAGGCCTGGGACGAGGGAAATATTGGACAGGTTGAGGTTAATCGAAGCGGAGGTACTT GCGAGGCCTGCCGAGGATGGTGATGTCAATGTGGGGAGCGTTAGGTTTATTTCGGGCGCCCGCAGGCCCGGGGCC GCACCTAGGATTCCGAGCTTTGGTATGGGAGTTGGGAAGGATATCAGAGGTGGCAGTGGTATGAGCAGTGCCAGT AGCGCCAGCTCCGATGAGAGTGACGAGGAGCTGATGGAGGCGGTTTCGAGGTTAACGATTACCAAGGCCGGATTG GAGGAAAGGTCGGCCTGGAGTACTTCATCTAGTGAGTAGGATTCCATCCCCTGCTTATTCGTCTGTTGACTAGTC GTACAGGTGTACAACCTCTACTCGACAATCAACATACGCCTAATGGATCAACCTATAGCACCTCCGTTATCCGTC AAGCTGCACATTCTAATACGCCCTCCGCACCCGCTCTATCCTCAATCCTGACTATCAGACCTTCACCCGATCCTA ATCGTGTCCCTGAACACGTCCTCCCGGCGACAACGTCCGACATATCTCACGCATCTACTGCGAGTGTGGACCCTC TGGAATCACCTTCGATCGTGTCCATCGCAACCCTCGACTCGTATCGCACAGCCAACACAGGTGAAAGCGTCATTT CAAGCACGTATGCAACTGAGGGTGGCTCCACCATTCGATCGCTGCATGCTCCTGGACATGCAGTCCTTCCGACTG TACATCGCTTCACATTGTTGAAGCCCGGTACAAAGCCGAAGAAGGTGTCGTTGGGTGGAGGACCGGGAGGTATAG GAGGGGCGGGAGGTACTGCTGGGGTGGTCACCCAGAGTTCTGTCGAAACCTCGGTCTGGAACCCGCTGGAGATAT TCTTCTCGAGCGGGTTATTGATTTCGAAATGTGACCTTTGTGCGAAGAGGATTGGGTGGAAGCCGTTTTTGGAGT GCGATGATTGTGGGTTGAGGTGTGTGTTTTTTCTCTTGTCTGGTCATCTCGGTATTGATTGTGTTGCGCGTAGGA CCCACGTCAAGTGCGGTGATCTGGCCCCGAGGGATTGTGGGATGCGAGCAGTTATGAATGGGCATCAACATTCGA CGCCTGTTGAGATGAAGAAGATTTCTCAGTCGGATTCGAAGTAGTACATATAATGTAGTTGTAGATGCAATTGGA CTAGTAATGTGTA |
| Length | 2113 |