CopciAB_263750
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_263750 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 263750 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_4:2987934..2990160 | Strand | + |
| Gene length (nt) | 2227 | Transcript length (nt) | 2163 |
| CDS length (nt) | 1263 | Protein length (aa) | 420 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_196107 | 73.5 | 3.62E-192 | 597 |
| Agrocybe aegerita | Agrae_CAA7261863 | 74 | 1.135E-191 | 596 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_116790 | 73 | 4.123E-191 | 594 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB16681 | 71.1 | 4.416E-186 | 580 |
| Schizophyllum commune H4-8 | Schco3_69382 | 65.5 | 2.967E-172 | 540 |
| Flammulina velutipes | Flave_chr07AA00199 | 62.1 | 1.804E-157 | 497 |
| Lentinula edodes B17 | Lened_B_1_1_9212 | 63.2 | 2.001E-154 | 488 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4952 | 63.2 | 2.509E-154 | 488 |
| Lentinula edodes NBRC 111202 | Lenedo1_1042002 | 65.2 | 2.754E-153 | 485 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1584789 | 58 | 6.153E-153 | 484 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_150064 | 60.5 | 4.215E-147 | 467 |
| Grifola frondosa | Grifr_OBZ70876 | 63.2 | 6.67E-132 | 423 |
| Pleurotus ostreatus PC9 | PleosPC9_1_84351 | 53.3 | 6.216E-112 | 365 |
| Auricularia subglabra | Aurde3_1_1161521 | 52.1 | 1.765E-84 | 286 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1025103 | 81.3 | 1.137E-81 | 277 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_263750.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd00180 | PKc | - | 191 | 377 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 181 | 363 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| D | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
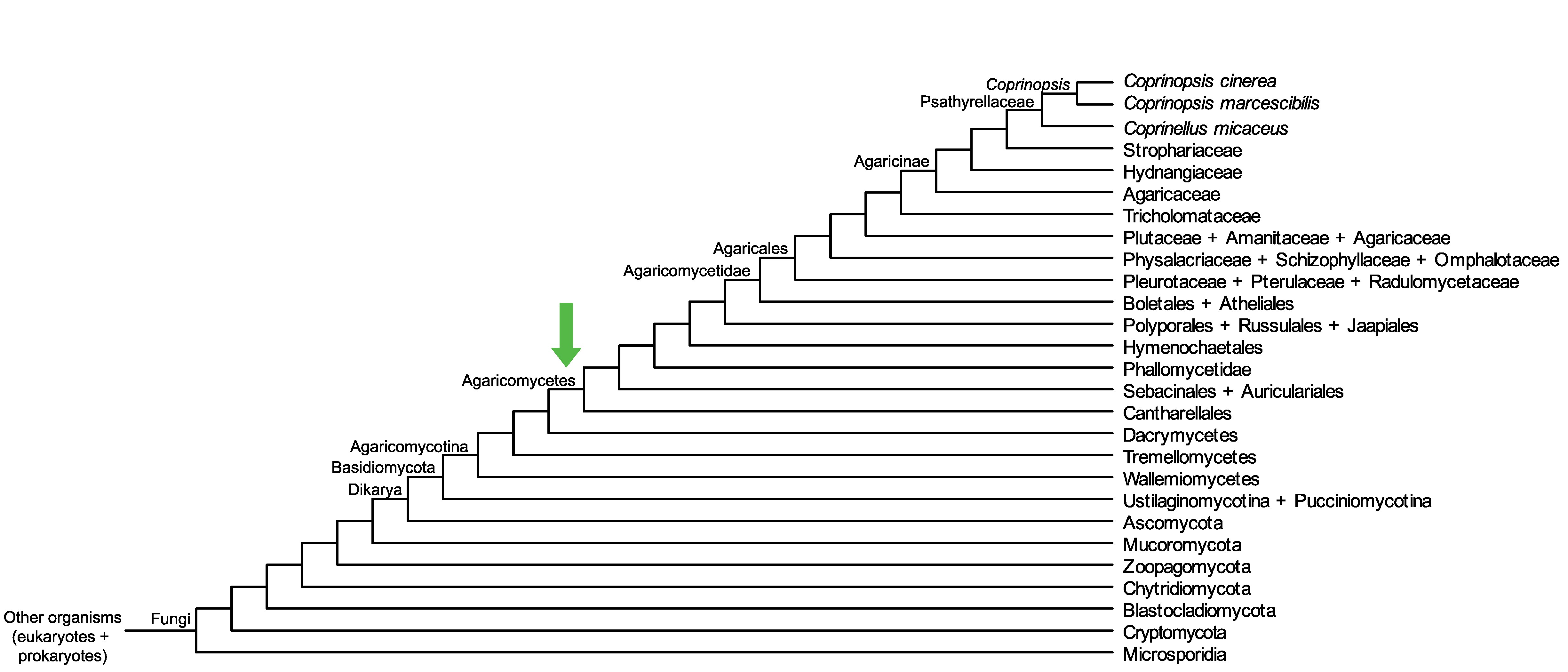
Conservation of CopciAB_263750 across fungi.
Arrow shows the origin of gene family containing CopciAB_263750.
Protein
| Sequence id | CopciAB_263750.T0 |
|---|---|
| Sequence |
>CopciAB_263750.T0 MAHRPGFGRNISVQVASPTNDLKAGVVSATTGPPSARTKPMQVKRVTLTSKKNWWMVEMDEEDEVQPLNNSINSF SSSEETAVEHEPAVVYKEYNHGIDEQVVHHHHHPSHKVEESPPPSPVYHPSRHPNSIFSFPTPTAYAAPPNATAS LTPFAHGSASELFKCVVKKPDVIQLEINGEMQSNASDVTVQFLAELRVYTTVARHRNICAFYGCLENVGIVLEYL ECRTLYDVIMARPPLTLAQKIDYHNQLLDGLTHLHSYRLSHGDLSLLNVQVTRDSNTIKLLDFGRSVSADSDFKS PHDEPVDPFPYLHSRQQPKLPAVRIEQIHPGTRPFSAPEILRGECQDPLLADAYSFGMILVCLDRCECVDVKPWD QRKDKLPGDLFENCELFEERAREYLKKWNEGRRALDKKDMIPEEI |
| Length | 420 |
Coding
| Sequence id | CopciAB_263750.T0 |
|---|---|
| Sequence |
>CopciAB_263750.T0 ATGGCACACCGACCAGGATTCGGCAGGAATATCAGCGTGCAGGTCGCCTCGCCGACCAACGACCTCAAGGCAGGC GTTGTATCTGCAACCACAGGGCCGCCGTCAGCTCGAACGAAACCGATGCAAGTGAAGCGGGTGACATTGACATCG AAGAAGAATTGGTGGATGGTGGAAATGGACGAGGAGGACGAAGTCCAGCCTCTGAACAACTCAATAAACTCTTTC TCCTCATCGGAAGAAACAGCCGTCGAACATGAACCAGCCGTTGTCTACAAGGAATACAACCATGGAATCGACGAG CAGGTCGTCCACCACCACCACCATCCATCCCACAAGGTCGAGGAATCGCCACCACCATCGCCAGTATATCACCCT TCGCGGCACCCCAATTCCATCTTCTCGTTCCCAACACCGACGGCCTACGCAGCGCCCCCCAATGCGACGGCCAGT TTAACACCATTCGCGCACGGATCAGCATCAGAATTGTTCAAATGTGTGGTGAAGAAGCCGGATGTCATTCAGCTC GAAATCAATGGCGAGATGCAGTCGAATGCTTCAGATGTGACCGTCCAGTTCCTGGCTGAACTGCGGGTGTACACA ACCGTCGCTCGACATCGCAATATCTGCGCTTTCTATGGCTGCTTGGAGAACGTCGGGATCGTGCTGGAGTATCTC GAATGCCGGACGTTATACGATGTGATAATGGCCCGGCCTCCCTTAACCCTCGCTCAAAAGATCGACTATCACAAT CAACTCCTCGACGGACTCACACACCTCCATTCATATCGGCTAAGTCACGGTGACTTGTCGTTACTGAACGTCCAG GTCACAAGGGACTCCAACACCATCAAGTTGCTCGACTTTGGGCGTTCGGTTTCTGCGGATTCGGACTTCAAATCG CCTCATGATGAACCCGTCGATCCGTTCCCCTACCTCCACAGCCGCCAGCAACCCAAACTTCCCGCCGTCAGGATC GAACAGATCCACCCCGGCACCCGACCTTTCTCCGCCCCGGAGATCCTGCGAGGCGAATGCCAGGATCCTCTGCTA GCCGATGCATATTCGTTTGGGATGATCCTTGTATGTCTGGATCGGTGCGAGTGTGTAGATGTGAAACCTTGGGAT CAGAGGAAGGACAAGCTACCGGGCGACCTGTTCGAAAATTGCGAACTGTTCGAGGAGAGGGCGAGGGAGTACCTG AAGAAGTGGAATGAAGGCAGACGGGCGTTGGACAAGAAGGATATGATACCGGAGGAAATTTGA |
| Length | 1263 |
Transcript
| Sequence id | CopciAB_263750.T0 |
|---|---|
| Sequence |
>CopciAB_263750.T0 GGCACTTTTGACTTGTACCATCTACCGCGCAAAGCGCAAACCTCGATTTATCGCCTCAGTTCTTATTTCTACCGC TTGATTCAACTTCTCAAATGGCACACCGACCAGGATTCGGCAGGAATATCAGCGTGCAGGTCGCCTCGCCGACCA ACGACCTCAAGGCAGGCGTTGTATCTGCAACCACAGGGCCGCCGTCAGCTCGAACGAAACCGATGCAAGTGAAGC GGGTGACATTGACATCGAAGAAGAATTGGTGGATGGTGGAAATGGACGAGGAGGACGAAGTCCAGCCTCTGAACA ACTCAATAAACTCTTTCTCCTCATCGGAAGAAACAGCCGTCGAACATGAACCAGCCGTTGTCTACAAGGAATACA ACCATGGAATCGACGAGCAGGTCGTCCACCACCACCACCATCCATCCCACAAGGTCGAGGAATCGCCACCACCAT CGCCAGTATATCACCCTTCGCGGCACCCCAATTCCATCTTCTCGTTCCCAACACCGACGGCCTACGCAGCGCCCC CCAATGCGACGGCCAGTTTAACACCATTCGCGCACGGATCAGCATCAGAATTGTTCAAATGTGTGGTGAAGAAGC CGGATGTCATTCAGCTCGAAATCAATGGCGAGATGCAGTCGAATGCTTCAGATGTGACCGTCCAGTTCCTGGCTG AACTGCGGGTGTACACAACCGTCGCTCGACATCGCAATATCTGCGCTTTCTATGGCTGCTTGGAGAACGTCGGGA TCGTGCTGGAGTATCTCGAATGCCGGACGTTATACGATGTGATAATGGCCCGGCCTCCCTTAACCCTCGCTCAAA AGATCGACTATCACAATCAACTCCTCGACGGACTCACACACCTCCATTCATATCGGCTAAGTCACGGTGACTTGT CGTTACTGAACGTCCAGGTCACAAGGGACTCCAACACCATCAAGTTGCTCGACTTTGGGCGTTCGGTTTCTGCGG ATTCGGACTTCAAATCGCCTCATGATGAACCCGTCGATCCGTTCCCCTACCTCCACAGCCGCCAGCAACCCAAAC TTCCCGCCGTCAGGATCGAACAGATCCACCCCGGCACCCGACCTTTCTCCGCCCCGGAGATCCTGCGAGGCGAAT GCCAGGATCCTCTGCTAGCCGATGCATATTCGTTTGGGATGATCCTTGTATGTCTGGATCGGTGCGAGTGTGTAG ATGTGAAACCTTGGGATCAGAGGAAGGACAAGCTACCGGGCGACCTGTTCGAAAATTGCGAACTGTTCGAGGAGA GGGCGAGGGAGTACCTGAAGAAGTGGAATGAAGGCAGACGGGCGTTGGACAAGAAGGATATGATACCGGAGGAAA TTTGAACACCTTCGCCGTTGACGGTTTTGGCCCGCCCGACTAGCCTGCTACTCCATCCTCATTCAGCCCCGCCTA TGCCGCCGCCTATCCATCTCCCCCTGATCCTGATCACAGCCCAACTACTGACCGCAGAAACCGTTTTCTACGGAC AAAAAGAGACGACAACGACGACGATGATGAAACCGGGGGTGCTTCCACGAACTCAGGAAAAGAAACATGACGACG ACCCTATGTTATATACACCCAACCCCTCCCCACTATGCCGCTCAATTTATAATCGCCTCATTCGCGAAGTACGCT ATTTCCGGAACCTTTTCGCGGAAATTATGCCATTTGTCGCTATACCGGAGTCCCGTCTGTGACCCTATTTAACCT CTTTCGAACTCTTTCGGTTTTTCTTGGTGGCCCTTTTTTTGTTCTCTCAAGTTTCGAACTTTCCGATTCTTTGGA ACGCTGTTGAGGGAAGGAAGGGCATATCGGGCTTGGCGGAGAGAAAGGAGAGGAGGATTGCTGCCGTGTTGGAAC CTGAAGGGTGCTTATCTGGAGAGGCTCATTGCAGACGGAGATTTCACTTGGTACAACCTCGTAATTATTCACTGA TCGTGTACTTCTTCCTATTGGACTTCCCCAGTCCGCTTACCTCTTGCTCATCATGGTTTGCTCTATTGTAGTCAT TTCATTCTTGTCGTCTGTTTCCTCGGTTCGTGTGCATAATTACTACCCCTTCTCTTCCCCGTGTAGCTACTAGTG TGACTGTTCTCTACTTTTTTGGAGCTGTTGTTACAATCGTGTAAATCACCTTGCTGGTGTTCA |
| Length | 2163 |
Gene
| Sequence id | CopciAB_263750.T0 |
|---|---|
| Sequence |
>CopciAB_263750.T0 GGCACTTTTGACTTGTACCATCTACCGCGCAAAGCGCAAACCTCGATTTATCGCCTCAGTTCTTATTTCTACCGC TTGATTCAACTTCTCAAATGGCACACCGACCAGGATTCGGCAGGAATATCAGCGTGCAGGTCGCCTCGCCGACCA ACGACCTCAAGGCAGGCGTTGTATCTGCAACCACAGGGCCGCCGTCAGCTCGAACGAAACCGATGCAAGTGAAGC GGGTGACATTGACATCGAAGAAGAATTGGTGGATGGGCGAGTTTAATCATCCTCTAGCCTTGAGACAAGCGCATG TCTAACCAAGCCTCGTAACATGCAGTGGAAATGGACGAGGAGGACGAAGTCCAGCCTCTGAACAACTCAATAAAC TCTTTCTCCTCATCGGAAGAAACAGCCGTCGAACATGAACCAGCCGTTGTCTACAAGGAATACAACCATGGAATC GACGAGCAGGTCGTCCACCACCACCACCATCCATCCCACAAGGTCGAGGAATCGCCACCACCATCGCCAGTATAT CACCCTTCGCGGCACCCCAATTCCATCTTCTCGTTCCCAACACCGACGGCCTACGCAGCGCCCCCCAATGCGACG GCCAGTTTAACACCATTCGCGCACGGATCAGCATCAGAATTGTTCAAATGTGTGGTGAAGAAGCCGGATGTCATT CAGCTCGAAATCAATGGCGAGATGCAGTCGAATGCTTCAGATGTGACCGTCCAGTTCCTGGCTGAACTGCGGGTG TACACAACCGTCGCTCGACATCGCAATATCTGCGCTTTCTATGGCTGCTTGGAGAACGTCGGGATCGTGCTGGAG TATCTCGAATGCCGGACGTTATACGATGTGATAATGGCCCGGCCTCCCTTAACCCTCGCTCAAAAGATCGACTAT CACAATCAACTCCTCGACGGACTCACACACCTCCATTCATATCGGCTAAGTCACGGTGACTTGTCGTTACTGAAC GTCCAGGTCACAAGGGACTCCAACACCATCAAGTTGCTCGACTTTGGGCGTTCGGTTTCTGCGGATTCGGACTTC AAATCGCCTCATGATGAACCCGTCGATCCGTTCCCCTACCTCCACAGCCGCCAGCAACCCAAACTTCCCGCCGTC AGGATCGAACAGATCCACCCCGGCACCCGACCTTTCTCCGCCCCGGAGATCCTGCGAGGCGAATGCCAGGATCCT CTGCTAGCCGATGCATATTCGTTTGGGATGATCCTTGTATGTCTGGATCGGTGCGAGTGTGTAGATGTGAAACCT TGGGATCAGAGGAAGGACAAGCTACCGGGCGACCTGTTCGAAAATTGCGAACTGTTCGAGGAGAGGGCGAGGGAG TACCTGAAGAAGTGGAATGAAGGCAGACGGGCGTTGGACAAGAAGGATATGATACCGGAGGAAATTTGAACACCT TCGCCGTTGACGGTTTTGGCCCGCCCGACTAGCCTGCTACTCCATCCTCATTCAGCCCCGCCTATGCCGCCGCCT ATCCATCTCCCCCTGATCCTGATCACAGCCCAACTACTGACCGCAGAAACCGTTTTCTACGGACAAAAAGAGACG ACAACGACGACGATGATGAAACCGGGGGTGCTTCCACGAACTCAGGAAAAGAAACATGACGACGACCCTATGTTA TATACACCCAACCCCTCCCCACTATGCCGCTCAATTTATAATCGCCTCATTCGCGAAGTACGCTATTTCCGGAAC CTTTTCGCGGAAATTATGCCATTTGTCGCTATACCGGAGTCCCGTCTGTGACCCTATTTAACCTCTTTCGAACTC TTTCGGTTTTTCTTGGTGGCCCTTTTTTTGTTCTCTCAAGTTTCGAACTTTCCGATTCTTTGGAACGCTGTTGAG GGAAGGAAGGGCATATCGGGCTTGGCGGAGAGAAAGGAGAGGAGGATTGCTGCCGTGTTGGAACCTGAAGGGTGC TTATCTGGAGAGGCTCATTGCAGACGGAGATTTCACTTGGTACAACCTCGTAATTATTCACTGATCGTGTACTTC TTCCTATTGGACTTCCCCAGTCCGCTTACCTCTTGCTCATCATGGTTTGCTCTATTGTAGTCATTTCATTCTTGT CGTCTGTTTCCTCGGTTCGTGTGCATAATTACTACCCCTTCTCTTCCCCGTGTAGCTACTAGTGTGACTGTTCTC TACTTTTTTGGAGCTGTTGTTACAATCGTGTAAATCACCTTGCTGGTGTTCA |
| Length | 2227 |