CopciAB_274375
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_274375 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 274375 |
| Uniprot id | Functional description | protein kinase activity | |
| Location | scaffold_8:1341453..1344783 | Strand | + |
| Gene length (nt) | 3331 | Transcript length (nt) | 2619 |
| CDS length (nt) | 2334 | Protein length (aa) | 777 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| 0 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_274375.T0 |
| Description | protein kinase activity |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd00882 | Ras_like_GTPase | - | 543 | 696 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 239 | 494 |
| Pfam | PF04548 | AIG1 family | IPR006703 | 542 | 719 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR006703 | AIG1-type guanine nucleotide-binding (G) domain |
| IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005525 | GTP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K17535 |
EggNOG
| COG category | Description |
|---|---|
| K | protein kinase activity |
| L | protein kinase activity |
| T | protein kinase activity |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
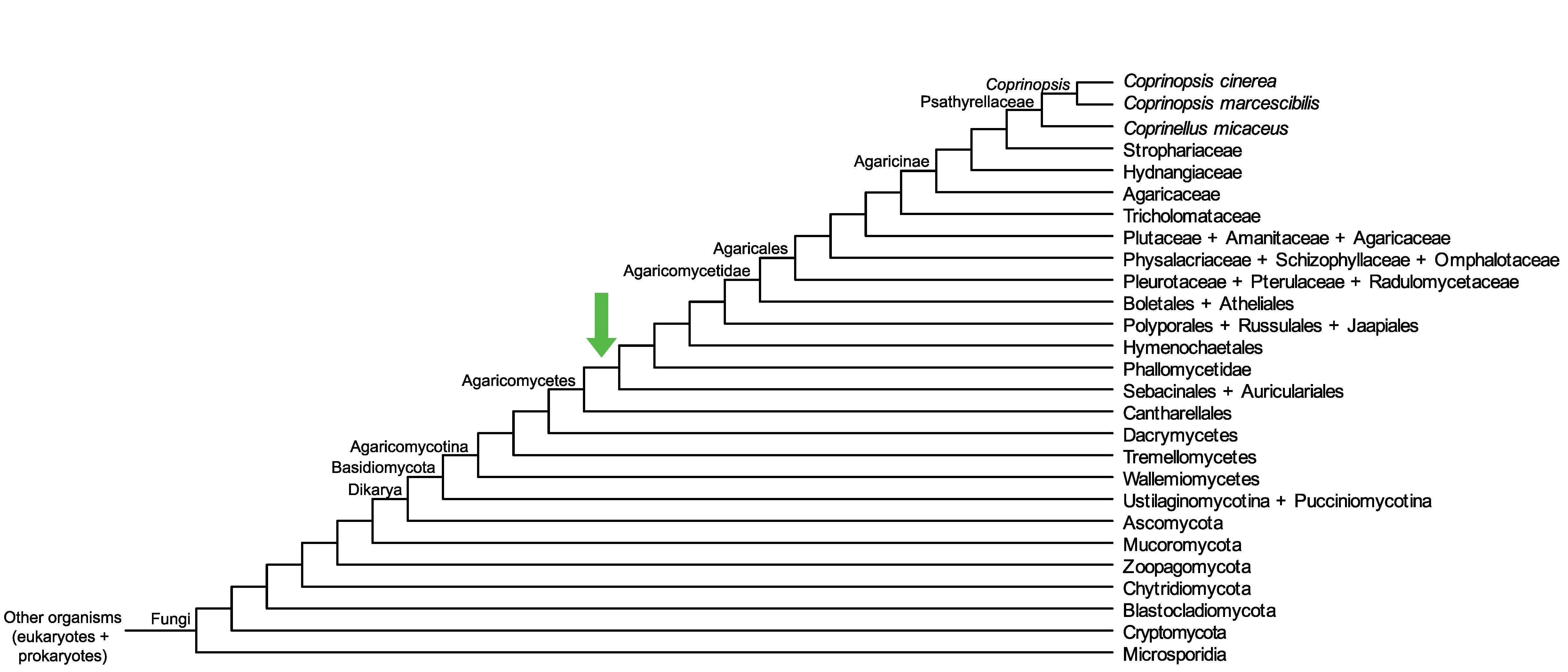
Conservation of CopciAB_274375 across fungi.
Arrow shows the origin of gene family containing CopciAB_274375.
Protein
| Sequence id | CopciAB_274375.T0 |
|---|---|
| Sequence |
>CopciAB_274375.T0 MPDSSLKSLAQQLASSSGLLTVSLLKADHRQWLERNMSYLHRVAADSRALADGVSPWPEAMLNLPFPRLQIFVRG CLCTIRGSQNRLLDVVTHEQTRWLELNARARLSRSLTIKHKLHQFIPYAAIQSVVRTDLVHRIIELDTANLAANV IAMCTDGEQRDAFLSAQDPAVDQELADLLYSVLGSRHLDPVHAPILRALLKLSENTDLLPQRLAHNSLQVAGGPP IHSTRISKIHRGKVDDRVVAVKVSNHSPELELGEYLNKAFKREAIIWSQLHHPNVLPFYGLFQAEGEHFYSLLSP WMDNGNITDFLKAEPQTDRQELIFDIARGLEYLHNLDPPVIHGDLKPNNIFVTANKRACIGDFGLGKLPRDPKWG ISRSSSLFAGNELYCAPELLEEIIEGEAESVSDATEDDQLGYKTLASDVYAFACICYEVYHGKPRFFDLHGLRRW KAIVGLIDDPTHKPGGMSDLLWHWVECFWDRNPAARPSISAFVQAMCPTPLNNPYKYDTAWDWTFLQDSPNDYDG SMKKVRPEELTDNDIVIALMGPTGTGKSTFINCALGDDVARVSGSVRSCTSKVELFACFHPDKPEQRVFFVDTPG FDHTEDVSSEKQTLAQVADWLIKTYKENIRLSGVLHFCSVTSARFSGTDRVNLRVFTELCGPEALDNVVIVSTGW EDLETDELVTLGEKREEQLRNTFLDDAGLDQVRYCRFSERTTQSAWRIIDLFQQQGNQRRPLLIQTEAVDKRLPL SQTSAFQAIRRRWNKWAKSWKGPKLLA |
| Length | 777 |
Coding
| Sequence id | CopciAB_274375.T0 |
|---|---|
| Sequence |
>CopciAB_274375.T0 ATGCCAGATTCATCGCTGAAGTCTCTGGCCCAACAATTGGCCTCCAGCTCTGGGCTTCTAACCGTATCACTGCTC AAGGCTGATCATAGGCAATGGCTGGAGCGGAATATGAGTTATCTCCACCGTGTGGCAGCAGATTCCAGGGCGTTG GCGGATGGTGTCAGCCCATGGCCAGAAGCAATGTTGAACTTACCCTTCCCACGTCTCCAGATATTCGTTCGAGGG TGCCTCTGCACGATACGCGGTAGCCAAAACAGACTTCTAGATGTAGTGACGCATGAGCAAACCCGGTGGCTGGAA CTGAATGCAAGAGCGCGATTATCCCGCTCTTTAACAATCAAGCACAAACTTCATCAGTTCATCCCATACGCAGCT ATTCAGTCGGTCGTCAGAACCGATCTGGTTCACCGCATTATAGAATTGGACACTGCGAACCTCGCCGCCAACGTG ATTGCCATGTGCACAGACGGGGAGCAACGCGACGCTTTCCTCAGCGCTCAGGATCCAGCTGTAGATCAGGAGCTC GCTGATCTACTCTACTCTGTACTTGGATCACGACATCTTGACCCAGTCCATGCACCCATCTTGCGCGCTCTCCTC AAGTTGTCGGAGAATACTGATCTCTTACCTCAACGCCTGGCCCACAACAGCCTGCAGGTTGCGGGTGGTCCACCC ATCCATAGCACTAGAATTAGCAAGATCCACAGAGGGAAGGTGGACGACCGTGTGGTGGCGGTCAAGGTTTCCAAC CATTCACCGGAGCTGGAGCTAGGGGAGTACTTGAACAAGGCGTTCAAGCGAGAAGCCATCATCTGGAGTCAGCTG CATCACCCTAACGTGCTGCCTTTCTACGGCCTGTTCCAGGCGGAGGGAGAACATTTTTACTCCCTCCTCTCACCT TGGATGGATAATGGAAACATCACGGACTTCCTCAAAGCGGAACCACAAACTGATCGACAAGAACTGATCTTCGAC ATCGCCAGAGGCCTAGAATACCTCCATAATCTTGATCCGCCTGTAATTCATGGGGATCTTAAACCGAATAATATC TTCGTCACTGCCAACAAGCGCGCTTGTATTGGCGACTTTGGTCTAGGCAAACTCCCTCGAGACCCCAAATGGGGC ATATCACGAAGTTCCAGCCTTTTTGCTGGGAATGAACTTTACTGCGCTCCAGAACTCCTCGAAGAGATTATCGAA GGAGAGGCGGAATCTGTCAGTGATGCCACAGAGGATGACCAACTGGGATACAAGACGCTTGCTAGTGATGTCTAT GCATTCGCCTGTATTTGTTACGAGGTTTATCATGGCAAACCTCGATTTTTTGATCTTCACGGCCTCAGGCGTTGG AAAGCAATAGTTGGGCTAATTGATGATCCAACCCACAAGCCTGGAGGGATGAGCGACTTGCTATGGCACTGGGTT GAATGCTTCTGGGATCGCAACCCTGCAGCAAGGCCATCAATTTCAGCGTTCGTGCAGGCAATGTGTCCGACTCCT CTCAACAATCCTTACAAGTACGACACGGCCTGGGATTGGACGTTCCTGCAAGACTCGCCTAATGACTATGATGGC TCAATGAAGAAGGTCAGACCTGAGGAATTGACCGACAATGACATTGTGATCGCATTGATGGGGCCTACTGGCACA GGGAAAAGCACTTTCATCAATTGCGCACTGGGTGACGATGTTGCTAGGGTTTCTGGCTCCGTCAGGTCGTGCACT TCGAAGGTCGAGCTGTTCGCGTGCTTCCACCCGGATAAACCAGAGCAACGAGTTTTCTTTGTGGACACTCCCGGT TTCGACCATACCGAAGACGTTAGCTCCGAGAAGCAGACGCTGGCACAAGTCGCAGATTGGCTGATAAAAACGTAC AAAGAAAACATCAGGCTCAGCGGTGTCCTCCACTTCTGTAGCGTCACTTCAGCACGTTTCTCAGGCACCGACCGA GTGAATCTCCGCGTTTTCACTGAACTATGCGGTCCGGAAGCTTTGGATAATGTCGTCATTGTATCGACTGGATGG GAAGACCTCGAGACTGATGAGCTAGTAACACTCGGCGAAAAGCGTGAAGAGCAATTGCGAAACACGTTTTTGGAT GACGCTGGCCTTGACCAAGTTCGCTACTGTCGCTTCTCGGAGAGGACAACGCAGAGCGCTTGGCGTATCATCGAC CTGTTCCAACAACAAGGCAATCAACGAAGGCCACTCCTGATACAAACAGAGGCCGTCGACAAGCGGCTACCGCTA TCGCAAACCTCTGCTTTTCAAGCCATTCGTAGAAGATGGAATAAATGGGCCAAATCTTGGAAAGGCCCAAAGCTC TTGGCTTAA |
| Length | 2334 |
Transcript
| Sequence id | CopciAB_274375.T0 |
|---|---|
| Sequence |
>CopciAB_274375.T0 ATCTAATGTCGTGGCCACGACTGAGCACCTCAATGATGGCCACGATAATCATCGCCAACTTCACTCCCTAGCGGA GCTTCGACACCGCGCGACCTGTTTCATTCCTTTCGTGATGCCAGATTCATCGCTGAAGTCTCTGGCCCAACAATT GGCCTCCAGCTCTGGGCTTCTAACCGTATCACTGCTCAAGGCTGATCATAGGCAATGGCTGGAGCGGAATATGAG TTATCTCCACCGTGTGGCAGCAGATTCCAGGGCGTTGGCGGATGGTGTCAGCCCATGGCCAGAAGCAATGTTGAA CTTACCCTTCCCACGTCTCCAGATATTCGTTCGAGGGTGCCTCTGCACGATACGCGGTAGCCAAAACAGACTTCT AGATGTAGTGACGCATGAGCAAACCCGGTGGCTGGAACTGAATGCAAGAGCGCGATTATCCCGCTCTTTAACAAT CAAGCACAAACTTCATCAGTTCATCCCATACGCAGCTATTCAGTCGGTCGTCAGAACCGATCTGGTTCACCGCAT TATAGAATTGGACACTGCGAACCTCGCCGCCAACGTGATTGCCATGTGCACAGACGGGGAGCAACGCGACGCTTT CCTCAGCGCTCAGGATCCAGCTGTAGATCAGGAGCTCGCTGATCTACTCTACTCTGTACTTGGATCACGACATCT TGACCCAGTCCATGCACCCATCTTGCGCGCTCTCCTCAAGTTGTCGGAGAATACTGATCTCTTACCTCAACGCCT GGCCCACAACAGCCTGCAGGTTGCGGGTGGTCCACCCATCCATAGCACTAGAATTAGCAAGATCCACAGAGGGAA GGTGGACGACCGTGTGGTGGCGGTCAAGGTTTCCAACCATTCACCGGAGCTGGAGCTAGGGGAGTACTTGAACAA GGCGTTCAAGCGAGAAGCCATCATCTGGAGTCAGCTGCATCACCCTAACGTGCTGCCTTTCTACGGCCTGTTCCA GGCGGAGGGAGAACATTTTTACTCCCTCCTCTCACCTTGGATGGATAATGGAAACATCACGGACTTCCTCAAAGC GGAACCACAAACTGATCGACAAGAACTGATCTTCGACATCGCCAGAGGCCTAGAATACCTCCATAATCTTGATCC GCCTGTAATTCATGGGGATCTTAAACCGAATAATATCTTCGTCACTGCCAACAAGCGCGCTTGTATTGGCGACTT TGGTCTAGGCAAACTCCCTCGAGACCCCAAATGGGGCATATCACGAAGTTCCAGCCTTTTTGCTGGGAATGAACT TTACTGCGCTCCAGAACTCCTCGAAGAGATTATCGAAGGAGAGGCGGAATCTGTCAGTGATGCCACAGAGGATGA CCAACTGGGATACAAGACGCTTGCTAGTGATGTCTATGCATTCGCCTGTATTTGTTACGAGGTTTATCATGGCAA ACCTCGATTTTTTGATCTTCACGGCCTCAGGCGTTGGAAAGCAATAGTTGGGCTAATTGATGATCCAACCCACAA GCCTGGAGGGATGAGCGACTTGCTATGGCACTGGGTTGAATGCTTCTGGGATCGCAACCCTGCAGCAAGGCCATC AATTTCAGCGTTCGTGCAGGCAATGTGTCCGACTCCTCTCAACAATCCTTACAAGTACGACACGGCCTGGGATTG GACGTTCCTGCAAGACTCGCCTAATGACTATGATGGCTCAATGAAGAAGGTCAGACCTGAGGAATTGACCGACAA TGACATTGTGATCGCATTGATGGGGCCTACTGGCACAGGGAAAAGCACTTTCATCAATTGCGCACTGGGTGACGA TGTTGCTAGGGTTTCTGGCTCCGTCAGGTCGTGCACTTCGAAGGTCGAGCTGTTCGCGTGCTTCCACCCGGATAA ACCAGAGCAACGAGTTTTCTTTGTGGACACTCCCGGTTTCGACCATACCGAAGACGTTAGCTCCGAGAAGCAGAC GCTGGCACAAGTCGCAGATTGGCTGATAAAAACGTACAAAGAAAACATCAGGCTCAGCGGTGTCCTCCACTTCTG TAGCGTCACTTCAGCACGTTTCTCAGGCACCGACCGAGTGAATCTCCGCGTTTTCACTGAACTATGCGGTCCGGA AGCTTTGGATAATGTCGTCATTGTATCGACTGGATGGGAAGACCTCGAGACTGATGAGCTAGTAACACTCGGCGA AAAGCGTGAAGAGCAATTGCGAAACACGTTTTTGGATGACGCTGGCCTTGACCAAGTTCGCTACTGTCGCTTCTC GGAGAGGACAACGCAGAGCGCTTGGCGTATCATCGACCTGTTCCAACAACAAGGCAATCAACGAAGGCCACTCCT GATACAAACAGAGGCCGTCGACAAGCGGCTACCGCTATCGCAAACCTCTGCTTTTCAAGCCATTCGTAGAAGATG GAATAAATGGGCCAAATCTTGGAAAGGCCCAAAGCTCTTGGCTTAATTTCGGACTGGAAATTGACCATCGTGACT CCTTTGGCTGATGTACGGATTCCATCATGTGTCCTTTGTACGACATTCTTATCACAATCGATCGCGTCTGGGAAC ATGCCTTGGTATGACATGGCCGCGTGGAAGCCTCGTTTCAACCAGCATGCTAGGAGCCAGCACTGCCTT |
| Length | 2619 |
Gene
| Sequence id | CopciAB_274375.T0 |
|---|---|
| Sequence |
>CopciAB_274375.T0 ATCTAATGTCGTGGCCACGACTGAGCACCTCAATGATGGCCACGATAATCATCGCCAACTTCACTCCCTAGCGGA GCTTCGACACCGCGCGACCTGTTTCATTCCTTTCGTGATGCCAGATTCATCGCTGAAGTCTCTGGCCCAACAATT GGCCTCCAGCTCTGGGCTTCTAACCGTATCACTGCTCAAGGCTGATCATAGGCAATGGCTGGAGCGGAATATGAG TTATCTCCACCGTGTGGCAGCAGATTCCAGGGCGTTGGCGGATGGTGTCAGCCCATGGCCAGAAGCAATGTTGAA CTTACCCTTCCCAGTTAATCCGCCTTCTGCCTCTTTCATATTTGTACCCCTAATATCTCACATCACAGCGTCTCC AGATATTCGTTCGAGGGTGCCTCTGCACGATACGCGGTAGCCAAAACAGGTAGGTCACACAAGCACAATCACTGT ATTGCATGTTGTGACCCTTTTGCAGACTTCTAGATGTAGTGACGCATGAGCAAACCCGGTGGCTGGAACTGAATG CAAGAGCGCGATTATCCCGCTCTTTAACAATCAAGCACAAACTTCATCAGTTCATCCCATACGCAGCTATTCAGG TAAGTGTTGCAAGGCTCGAGCAGAAGAGGTATTTGACTGACCGGAATTGCAAGTCGGTCGTCAGAACCGATCTGG TTCACCGCATTATAGAATTGGACACTGCGAACCTCGCCGCCAACGTGATTGCCATGTGCACAGACGGGGAGCAAC GCGACGCTTTCCTCAGCGCTCAGGATCCAGCTGTAGATCAGGAGCTCGCTGATCTACTCTACTCTGTTAGTCGAA TTGTTCGCTCTCTCTCGCTCCTCACCTCACCTGCTTCATTTAGGTACTTGGATCACGACATCTTGACCCAGTCCA TGCACCCATCTTGCGCGCTCTCCTCAAGTTGTCGGAGAATACTGATCTCTTACCTCAACGCCTGGCCCACAACAG CCTGCAGGTTGCGGGTGGTCCACCCATCCATAGCACTAGAATTAGCAAGATCCACAGAGGGAAGGTGGACGACCG TGTGGTGGCGGTCAAGGTTTCCAACCATTCACCGGAGCTGGAGCTAGGGGAGTACTTGAACAAGGTAGGTTATCT TCTATACGAAACTTGATCACCCCTGACATGTATCACCTTAGGCGTTCAAGCGAGAAGCCATCATCTGGAGTCAGC TGCATCACCCTAACGTGCTGCCTTTCTACGGCCTGTTCCAGGCGGAGGGAGAACATTTTTACTCCCTCCTCTCAC CTTGGATGGATAATGGAAACATCACGGACTTCCTCAAAGCGGAACCACAAACTGATCGACAAGAACTGGTTCGTT TTCGTTTACCGTTCGCCTTTCTCCTTGGACTCACGCGCGCGGTCTGTAAGATCTTCGACATCGCCAGAGGCCTAG AATACCTCCATAATCTTGATCCGCCTGTAATTCATGGGGATCTTAAACCGGTATGCCTGTTTATCGTCGCTGGAG CAATGTTCACTCACCGTCCACCGAAGAATAATATCTTCGTCACTGCCAACAAGCGCGCTTGTATTGGCGACTTTG GTCTAGGCAAACTCCCTCGAGACCCCAAATGGGGCATATCACGAAGTTCCAGCCTTTTTGCTGGGAATGAACTTT ACTGCGCTCCAGAACTCCTCGAAGAGATTATCGAAGGAGAGGCGGAATCTGTCAGTGATGCCACAGAGGATGACC AACTGGGATACAAGACGCTTGCTAGTGATGTCTATGCATTCGCCTGTATTTGTTACGAGGTGCGTAAGGTCCCTT TTTGCTTCAAGGATACCGCAAATGTATTCTTACCGTCCAGGTTTATCATGGCAAACCTCGATTTTTTGATCTTCA CGGCCTCAGGCGTTGGAAAGCAATAGTTGGGCTAATTGATGATCCAACCCACAAGCCTGGAGGGATGAGCGACTT GCTATGGCACTGGGTTGAATGCTTCTGGGATCGCAACCCTGCAGCAAGGCCATCAATTTCAGCGTTCGTGCAGGC AATGTGTCCGACTCCTCTCAACAATCCTTACAAGTACGACACGGCCTGGGATTGGACGTTCCTGCAAGGTGAGAT TCCAACGGGATGGGCTGTCAATTGAATCTGACCACTTGGCGTAGACTCGCCTAATGACTATGATGGCTCAATGAA GAAGGTCAGACCTGAGGAATTGACCGACAATGACATTGTGATCGCGTTGGTACTTACTGTTTATTCTTTGAACGT AGTTGCTGACTGTCTTCCAGATTGATGGGGCCTACTGGCACAGGGAAAAGCACTGTAAGTGTCCCGGCCGTCCTG TCTCTTCCGATTCGTCTTGCTGACTCCCGTCGCGACAGTTCATCAATTGCGCACTGGGTGACGATGTTGCTAGGG TTTCTGGCTCCGTCAGGTCGTGCACTTCGAAGGTCGAGCTGTTCGCGTGCTTCCACCCGGATAAACCAGAGCAAC GAGTTTTCTTTGTGGACACTCCCGGTTTCGACCATACCGAAGACGTTAGCTCCGAGAAGCAGACGCTGGCACAAG TCGCAGATTGGCTGATAAAAACGTACGTCATCCTTTTTGTTCCGCTGTGGATTGGGCTGCTGAGACAATTGCAGG TACAAAGAAAACATCAGGCTCAGCGGTGTCCTCCACTTCTGTAGCGTCACTTCAGCACGTTTCTCAGGCACCGAC CGAGTGAATCTCCGCGTTTTCACTGAACTATGCGGTCCGGAAGCTTTGGATAATGTCGTCATTGTATCGACTGGA TGGGAAGACCTCGAGACTGATGAGCTAGTAACACTCGGCGAAAAGCGTGAAGAGCAATTGCGAAACACGTTTTTG GATGACGCTGGCCTTGACCAAGTTCGCTACTGTCGCTTCTCGGAGAGGACAACGCAGAGCGCTTGGCGTATCATC GACCTGTTCCAACAACAAGGCAATCAACGAAGGCCACTCCTGATACAAACAGAGGCCGTCGACAAGCGGCTACCG CTATCGCAAACCTCTGCTTTTCAAGCCATTCGTAGAAGATGGAATAAATGGGCCAAATCTTGGAAAGGCCCAAAG CTCTTGGCTTAATTTCGGACTGGAAATTGACCATCGTGACTCCTTTGGCTGATGGTCAGTGAAACTCCGTCCCGT CGAAGTCCAGCCTTTACTTACCCCGCTTATCGTCCGTATCTCTTTGGCAGTACGGATTCCATCATGTGTCCTTTG TACGACATTCTTATCACAATCGATCGCGTCTGGGAACATGCCTTGGTATGACATGGCCGCGTGGAAGCCTCGTTT CAACCAGCATGCTAGGAGCCAGCACTGCCTT |
| Length | 3331 |