CopciAB_353965
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_353965 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 353965 |
| Uniprot id | Functional description | Glycosyltransferase family 15 protein | |
| Location | scaffold_13:503644..505801 | Strand | + |
| Gene length (nt) | 2158 | Transcript length (nt) | 1797 |
| CDS length (nt) | 1167 | Protein length (aa) | 388 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YDR483W_KRE2 |
| Candida albicans | MNT2 |
| Neurospora crassa | NCU06541 |
| Aspergillus nidulans | AN2752 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264528 | 67.9 | 5.519E-193 | 598 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21751 | 70.2 | 6.214E-193 | 598 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_211452 | 66.6 | 1.926E-192 | 596 |
| Schizophyllum commune H4-8 | Schco3_2629206 | 64.4 | 2.497E-185 | 576 |
| Pleurotus eryngii ATCC 90797 | Pleery1_187145 | 62.7 | 2.902E-182 | 567 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1054660 | 63.1 | 2.467E-182 | 567 |
| Pleurotus ostreatus PC9 | PleosPC9_1_115075 | 63.1 | 2.415E-182 | 567 |
| Lentinula edodes B17 | Lened_B_1_1_628 | 65 | 2.725E-180 | 561 |
| Lentinula edodes NBRC 111202 | Lenedo1_1043354 | 65 | 3.444E-180 | 561 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_12705 | 64.7 | 3.737E-179 | 558 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_55798 | 62.3 | 8.389E-178 | 554 |
| Flammulina velutipes | Flave_chr05AA00885 | 70.2 | 2.451E-165 | 518 |
| Grifola frondosa | Grifr_OBZ72892 | 56.1 | 1.706E-160 | 504 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_131474 | 59.1 | 5.388E-154 | 485 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_353965.T0 |
| Description | Glycosyltransferase family 15 protein |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF01793 | Glycolipid 2-alpha-mannosyltransferase | IPR002685 | 57 | 344 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 23 | 0.7135 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 7 | 29 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR002685 | Glycosyl transferase, family 15 |
| IPR029044 | Nucleotide-diphospho-sugar transferases |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0000030 | mannosyltransferase activity | MF |
| GO:0006486 | protein glycosylation | BP |
| GO:0016020 | membrane | CC |
KEGG
| KEGG Orthology |
|---|
| K10967 |
EggNOG
| COG category | Description |
|---|---|
| G | Glycosyltransferase family 15 protein |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GT | GT15 |
Transcription factor
| Group |
|---|
| No records |
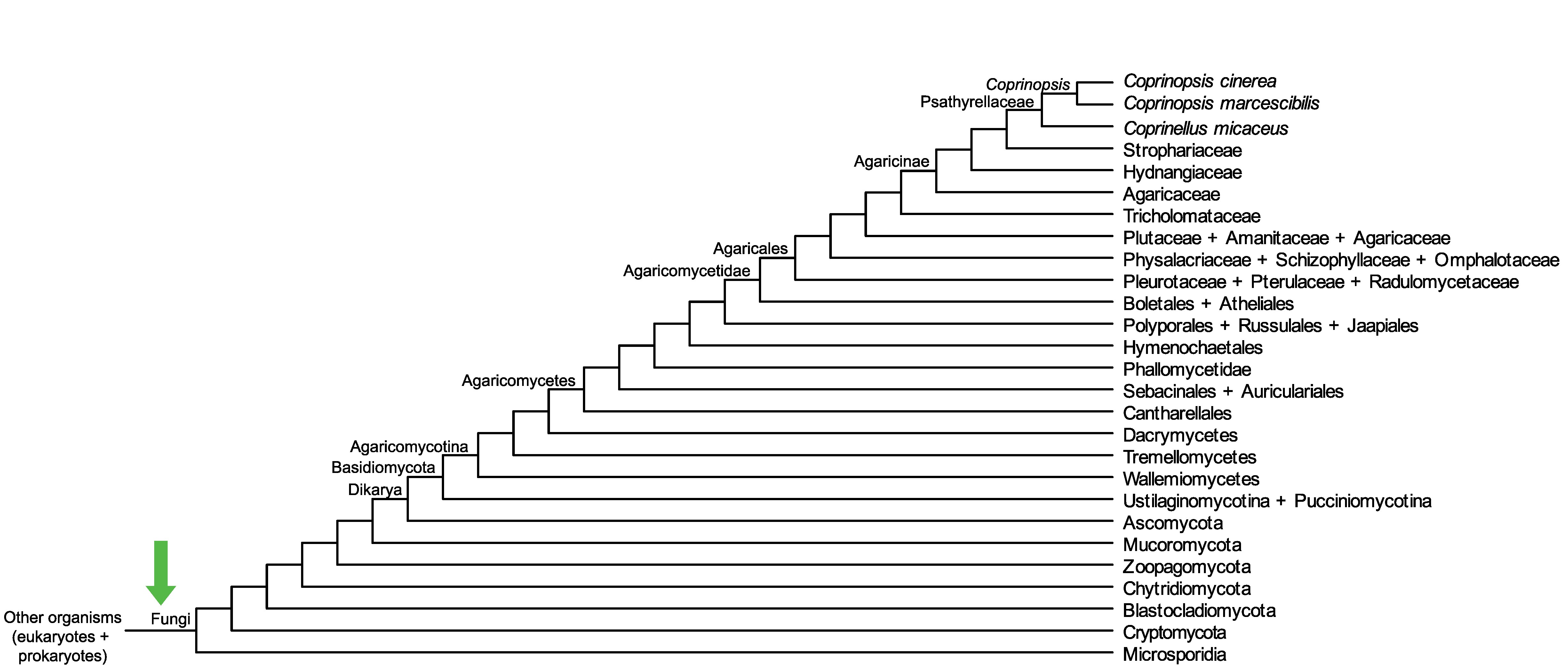
Conservation of CopciAB_353965 across fungi.
Arrow shows the origin of gene family containing CopciAB_353965.
Protein
| Sequence id | CopciAB_353965.T0 |
|---|---|
| Sequence |
>CopciAB_353965.T0 MNTPTRYILLALFLIISLHYLLAFTHEAYGRATALDHLKDKIVGPAPKPHVDLDTLYGPDVLTGDAPLKQDKKAN ATFVILCRNSDMGNIVRSVREIEDRFNRYHRYPYVFLNEEPFSEEFKMRLSVLSAAKMEFGQIPRDHWYQPDWID EEKAKASREKMVKEKVIYGGSVSYRNMCRFNSGFFFRHPLMLKYRWYWRIEPDVHFHCNINHDPFLYMQEHNKTY GFTITLYEYERTIPTLWGHVQDFMKENPQYVAEDNALGFMSERNGEKYNLCHFWSNFEIADMEFWRSPAYMDFFN YLDSKGGFYYERWGDAPVHSIAAAIFLNKNQIHFFDEIGYEHAPYTHCPKLGKNWENGRCSCDQKKSFDYDGYSC MRKWDAFMGLPSH |
| Length | 388 |
Coding
| Sequence id | CopciAB_353965.T0 |
|---|---|
| Sequence |
>CopciAB_353965.T0 ATGAATACCCCGACACGATACATCCTTCTCGCATTATTCTTAATCATCTCGCTGCACTACCTCCTCGCCTTTACA CACGAGGCATACGGACGTGCTACGGCGCTCGACCATCTCAAGGACAAGATCGTCGGGCCAGCGCCGAAACCCCAT GTCGACCTCGATACGCTGTACGGACCTGACGTCCTGACTGGCGACGCACCCCTGAAGCAGGACAAGAAGGCCAAC GCGACGTTTGTCATTCTGTGCCGGAACAGCGATATGGGCAACATCGTGCGCAGCGTGCGGGAGATTGAGGATCGG TTCAACCGGTACCACCGATATCCTTATGTTTTCCTCAACGAAGAACCCTTCTCTGAAGAATTCAAGATGCGACTG AGCGTGCTGAGCGCTGCCAAGATGGAGTTTGGCCAGATCCCGCGCGACCACTGGTACCAACCCGACTGGATCGAC GAAGAGAAAGCCAAAGCCAGCCGCGAGAAGATGGTCAAGGAGAAGGTTATCTACGGTGGTAGCGTCTCGTACCGC AACATGTGCCGATTCAACTCTGGGTTCTTCTTCCGCCACCCGTTGATGCTTAAATACCGGTGGTACTGGCGTATC GAGCCTGATGTCCACTTCCACTGCAATATCAACCACGATCCCTTCCTCTACATGCAGGAGCACAACAAGACCTAT GGATTCACAATCACGTTGTACGAGTACGAGAGGACTATTCCGACGCTTTGGGGACACGTCCAAGACTTTATGAAG GAAAACCCTCAATACGTCGCCGAGGACAACGCACTCGGGTTCATGTCAGAACGCAACGGCGAGAAATACAACCTC TGCCATTTCTGGTCCAACTTTGAAATTGCCGACATGGAATTCTGGCGCAGCCCCGCCTATATGGACTTTTTCAAC TACCTCGACTCCAAAGGTGGGTTCTACTACGAACGCTGGGGTGACGCGCCCGTGCACAGCATCGCCGCCGCCATC TTCCTCAACAAGAACCAGATCCACTTTTTCGATGAGATTGGGTACGAGCATGCGCCGTATACGCATTGTCCCAAG CTCGGAAAGAATTGGGAGAATGGGAGGTGCTCGTGTGATCAGAAGAAGAGCTTTGATTATGATGGATATTCGTGC ATGAGGAAGTGGGATGCTTTCATGGGCTTGCCTAGCCATTAG |
| Length | 1167 |
Transcript
| Sequence id | CopciAB_353965.T0 |
|---|---|
| Sequence |
>CopciAB_353965.T0 ATTTTCTCCTTTTCTCCTCCAAGAGCCTGTTACGTTCCAACCGTCCTCTGACCCCTCCTCGACAGCCCCAGGAGC CCACAGAGCGTCGAGGACCCACTTTCTCCCGGGGAAATCCATCCTTTTAAAAATTGCAACGCGATCGCCGACACG CGCCACACCACAAATCGCAATTTAGCAGCGGTCAGCAGGTGGTTTGCTGGCAGCCATAAACCTCGGGCGCGCGAC ATAGAAACATCGACTTGACACCCTCTATAGAACCAAGATGAATACCCCGACACGATACATCCTTCTCGCATTATT CTTAATCATCTCGCTGCACTACCTCCTCGCCTTTACACACGAGGCATACGGACGTGCTACGGCGCTCGACCATCT CAAGGACAAGATCGTCGGGCCAGCGCCGAAACCCCATGTCGACCTCGATACGCTGTACGGACCTGACGTCCTGAC TGGCGACGCACCCCTGAAGCAGGACAAGAAGGCCAACGCGACGTTTGTCATTCTGTGCCGGAACAGCGATATGGG CAACATCGTGCGCAGCGTGCGGGAGATTGAGGATCGGTTCAACCGGTACCACCGATATCCTTATGTTTTCCTCAA CGAAGAACCCTTCTCTGAAGAATTCAAGATGCGACTGAGCGTGCTGAGCGCTGCCAAGATGGAGTTTGGCCAGAT CCCGCGCGACCACTGGTACCAACCCGACTGGATCGACGAAGAGAAAGCCAAAGCCAGCCGCGAGAAGATGGTCAA GGAGAAGGTTATCTACGGTGGTAGCGTCTCGTACCGCAACATGTGCCGATTCAACTCTGGGTTCTTCTTCCGCCA CCCGTTGATGCTTAAATACCGGTGGTACTGGCGTATCGAGCCTGATGTCCACTTCCACTGCAATATCAACCACGA TCCCTTCCTCTACATGCAGGAGCACAACAAGACCTATGGATTCACAATCACGTTGTACGAGTACGAGAGGACTAT TCCGACGCTTTGGGGACACGTCCAAGACTTTATGAAGGAAAACCCTCAATACGTCGCCGAGGACAACGCACTCGG GTTCATGTCAGAACGCAACGGCGAGAAATACAACCTCTGCCATTTCTGGTCCAACTTTGAAATTGCCGACATGGA ATTCTGGCGCAGCCCCGCCTATATGGACTTTTTCAACTACCTCGACTCCAAAGGTGGGTTCTACTACGAACGCTG GGGTGACGCGCCCGTGCACAGCATCGCCGCCGCCATCTTCCTCAACAAGAACCAGATCCACTTTTTCGATGAGAT TGGGTACGAGCATGCGCCGTATACGCATTGTCCCAAGCTCGGAAAGAATTGGGAGAATGGGAGGTGCTCGTGTGA TCAGAAGAAGAGCTTTGATTATGATGGATATTCGTGCATGAGGAAGTGGGATGCTTTCATGGGCTTGCCTAGCCA TTAGACCTTTTCGTTCGGGTTTGCACGCAGGGGGTAGTGGAGGCCATGTCACGGGAATTCAAGTATCTCTTCTTT GCTCAAGTAGTTCGGTTGGAGAAATCTGCGATTGCATATCTTTCGACGAGTTCTGTCCACTGCTGGGACATACCA CAGCGTGCTATTACCATAACCACATTCAGGACCGTCTTTTCGTTCTTTGTCTCGGGCCTCTCCACCCTATATATC TACTTTGATCATAAGCTTCTTCGACTGGACCCCTACCCTTGAAACATCCCTTCTACATACTTCTGCTGTACTTAC ACTTCCCCTCCCTTGCTATATTGGTTTTAATCGTGGATTGTAAAACACTTGTAATAGATCTGGTCTGCTGTC |
| Length | 1797 |
Gene
| Sequence id | CopciAB_353965.T0 |
|---|---|
| Sequence |
>CopciAB_353965.T0 ATTTTCTCCTTTTCTCCTCCAAGAGCCTGTTACGTTCCAACCGTCCTCTGACCCCTCCTCGACAGCCCCAGGAGC CCACAGAGCGTCGAGGACCCACTTTCTCCCGGGGAAATCCATCCTTTTAAAAATTGCAACGCGATCGCCGACACG CGCCACACCACAAATCGCAATTTAGCAGCGGTCAGCAGGTGGTTTGCTGGCAGCCATAAACCTCGGGCGCGCGAC ATAGAAACATCGACTTGACACCCTCTATAGAACCAAGATGAATACCCCGACACGATACATCCTTCTCGCATTATT CTTAATCATCTCGCTGCACTACCTCCTCGCCTTTACACACGAGGCATACGGACGTGCTACGGCGCTCGACCATCT CAAGGACAAGATCGTCGGGCCAGCGCCGAAACCCCATGTCGACCTCGATACGCTGTACGGACCTGACGTCCTGAC TGGCGACGCACCCCTGAAGCAGGACAAGAAGGCCAACGCGACGTTTGTCATTCTGTGCCGGAACAGCGATATGGG CAACATCGTGCGCAGCGTGCGGGAGATTGAGGATCGGTTCAACCGGTACCACCGATATCCTTATGTTTTCCTCAA CGAAGAACCCTTCTCTGAAGAATTCAAGATGTGAGTAATCGATTCCTTTTTTCTTATGGGCGGTGACTGATTTCC GAACGAACGAAATCCCAGGCGACTGAGCGTGCTGAGCGCTGCCAAGATGGAGTTTGGCCAGATCCCGCGCGACCA CTGGTACCAACCCGACTGGATCGACGAAGAGAAAGCCAAAGCCAGCCGCGAGAAGATGGTCAAGGAGAAGGTTAT CTACGGTGGTAGCGTCTCGTACCGCAACATGTGCCGATTCAACTCTGGGTTCTTCTTCCGCCACCCGTTGATGCT TAAATACCGGTGGTACTGGCGTATCGAGTGGGTATTTTTATGTTCATGTTCATGGTTGTGGTTTGTGAGGTTAGG ATGAGTGCTGATGGGATGCGTTGTTATAGGCCTGATGTCCACTTCCACTGCAATATCAACCACGATCCCTTCCTC TACATGCAGGAGCACAACAAGACCTATGGATTCACAATCACGTTGTACGAGTACGAGAGGACTATTCCGACGCTT TGGGGACACGTCCAAGGTTCGTCTATACCCCCGTTTCAGCGTCCCGTCGATTCTAAACCTCCTCTTCAGACTTTA TGAAGGAAAACCCTCAATACGTCGCCGAGGACAACGCACTCGGGTTCATGTCAGAACGCAACGGCGAGAAATACA ACCTCTGCCATTGTACGTCTCCTCTCCCTCTCCCTCTCCTCCCTCCCTCTCCTTCTCCTAACCTTAAACTTAAAA CTAAACCCACACACCCGTAAACAGTCTGGTCCAACTTTGAAATTGCCGACATGGAATTCTGGCGCAGCCCCGCCT ATATGGACTTTTTCAACTACCTCGACTCCAAAGGTGGGTTCTACTACGAACGCTGGGGTGACGCGCCCGTGCACA GCATCGCCGCCGCCATCTTCCTCAACAAGAACCAGATCCACTTTTTCGATGAGATTGGGTACGAGCATGCGCCGT ATACGCATTGTCCCAAGCTCGGAAAGAATTGGGAGAATGGGAGGTGCTCGTGTGATCAGAAGAAGAGCTTTGGTG AGTGGTTTTTTCTTTTGGGTTGTGGTTTTGGTTTGGTGTGAGGTGAGATGGGATGCTGATGGTTGTGCGTTTTGG TAGATTATGATGGATATTCGTGCATGAGGAAGTGGGATGCTTTCATGGGCTTGCCTAGCCATTAGACCTTTTCGT TCGGGTTTGCACGCAGGGGGTAGTGGAGGCCATGTCACGGGAATTCAAGTATCTCTTCTTTGCTCAAGTAGTTCG GTTGGAGAAATCTGCGATTGCATATCTTTCGACGAGTTCTGTCCACTGCTGGGACATACCACAGCGTGCTATTAC CATAACCACATTCAGGACCGTCTTTTCGTTCTTTGTCTCGGGCCTCTCCACCCTATATATCTACTTTGATCATAA GCTTCTTCGACTGGACCCCTACCCTTGAAACATCCCTTCTACATACTTCTGCTGTACTTACACTTCCCCTCCCTT GCTATATTGGTTTTAATCGTGGATTGTAAAACACTTGTAATAGATCTGGTCTGCTGTC |
| Length | 2158 |