CopciAB_354384
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_354384 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | CKA1 | Synonyms | 354384 |
| Uniprot id | Functional description | Belongs to the protein kinase superfamily | |
| Location | scaffold_1:3470341..3472047 | Strand | - |
| Gene length (nt) | 1707 | Transcript length (nt) | 1321 |
| CDS length (nt) | 1044 | Protein length (aa) | 347 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YIL035C_CKA1 |
| Schizosaccharomyces pombe | cka1 |
| Aspergillus nidulans | AN1485_cka1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29144 | 91.9 | 1.356E-224 | 687 |
| Agrocybe aegerita | Agrae_CAA7258782 | 91.3 | 1.589E-221 | 678 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1535271 | 89.8 | 2.216E-217 | 666 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1092161 | 89.8 | 1.884E-217 | 666 |
| Pleurotus ostreatus PC9 | PleosPC9_1_85419 | 89.8 | 1.844E-217 | 666 |
| Schizophyllum commune H4-8 | Schco3_2612505 | 90.5 | 2.737E-216 | 663 |
| Lentinula edodes NBRC 111202 | Lenedo1_251252 | 84.7 | 6.442E-214 | 656 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_189988 | 91.7 | 1.059E-213 | 655 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_110117 | 91.7 | 1.104E-213 | 655 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_15404 | 87 | 2.251E-208 | 640 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_113270 | 86.7 | 4.624E-208 | 639 |
| Auricularia subglabra | Aurde3_1_1228088 | 84.9 | 5.305E-204 | 628 |
| Grifola frondosa | Grifr_OBZ79038 | 82.9 | 6.858E-195 | 601 |
| Lentinula edodes B17 | Lened_B_1_1_4209 | 93.2 | 4.286E-109 | 353 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | CKA1 |
|---|---|
| Protein id | CopciAB_354384.T0 |
| Description | Belongs to the protein kinase superfamily |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14132 | STKc_CK2_alpha | IPR045216 | 21 | 326 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 40 | 325 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017441 | Protein kinase, ATP binding site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR045216 | Casein Kinase 2, subunit alpha |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K03097 |
EggNOG
| COG category | Description |
|---|---|
| D | Belongs to the protein kinase superfamily |
| K | Belongs to the protein kinase superfamily |
| T | Belongs to the protein kinase superfamily |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
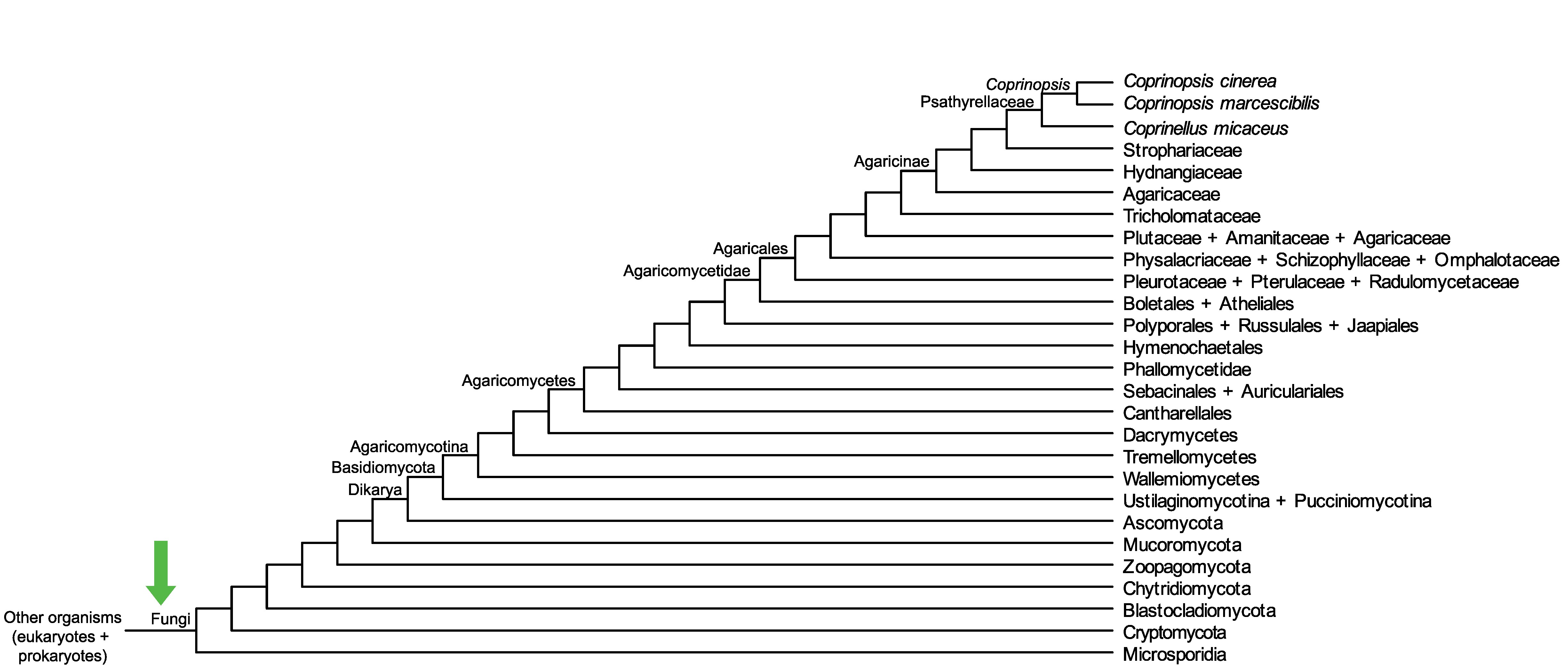
Conservation of CopciAB_354384 across fungi.
Arrow shows the origin of gene family containing CopciAB_354384.
Protein
| Sequence id | CopciAB_354384.T0 |
|---|---|
| Sequence |
>CopciAB_354384.T0 MGRSTTKSVARVYADVNARLGPSWHEYDNLQVQWGSQDHYEIVRKVGRGKYSEVFEGINVVTEEKCIIKVLKPVK KKKIKREIKILQNLAGGPNIVALLDVVRDPASKIPSLITEYVHNVEFKQLYPRFTDMDVRYYMFELLKALDFCHS KGIMHRDVKPHNVMIDHEHRKLRLIDWGLAEFYHPKTEYNVRVASRYFKGPELLVDFQEYDYSLDMWSYGCMFAS MIFRKEPFFHGHDNYDQLVKITKVLGTDDLYAYIEKYNIRLDSQYDEILGRYPRKPWTRFITSENQRYISNEAID FLDKLLRYDHQDRLTAREAQMQPYFDPIRNAATNNAHAGESDDASSG |
| Length | 347 |
Coding
| Sequence id | CopciAB_354384.T0 |
|---|---|
| Sequence |
>CopciAB_354384.T0 ATGGGGCGCTCAACAACAAAATCGGTCGCCCGGGTGTACGCTGATGTCAACGCTCGCTTAGGGCCTTCATGGCAT GAATACGACAACCTCCAAGTTCAATGGGGATCTCAGGATCATTATGAAATCGTGCGGAAAGTGGGAAGGGGAAAG TACTCAGAGGTTTTCGAAGGAATCAATGTTGTAACGGAAGAGAAATGTATTATTAAAGTCCTCAAGCCTGTCAAG AAGAAGAAAATCAAGCGAGAAATCAAGATCCTTCAAAACTTGGCCGGAGGTCCCAATATCGTCGCTTTGCTCGAT GTCGTCCGCGACCCTGCATCCAAGATCCCCAGCCTGATAACAGAATACGTGCACAACGTCGAGTTCAAGCAATTA TACCCCCGATTCACCGACATGGACGTTCGCTACTACATGTTTGAACTCCTTAAGGCTCTGGACTTCTGTCACTCC AAAGGAATCATGCACCGCGACGTTAAGCCGCACAACGTGATGATTGATCATGAGCACCGTAAATTGCGCTTGATC GACTGGGGTCTGGCTGAATTCTACCATCCAAAGACGGAATACAACGTTCGCGTGGCCTCTCGCTACTTCAAGGGA CCTGAGCTCTTGGTCGACTTCCAAGAGTATGACTACAGCCTGGATATGTGGAGCTATGGTTGCATGTTCGCCTCG ATGATCTTCCGCAAGGAACCCTTCTTCCACGGACACGACAACTATGACCAACTGGTTAAAATCACCAAAGTCCTT GGTACAGACGACCTCTACGCCTATATCGAGAAGTACAACATCCGCCTTGACTCTCAATACGACGAGATTCTCGGA AGGTATCCTCGCAAGCCCTGGACCCGCTTCATCACCTCGGAAAACCAGAGATACATCTCCAACGAGGCCATCGAT TTCCTTGACAAGCTCCTCCGTTACGACCACCAAGACCGTCTCACTGCTCGTGAGGCCCAGATGCAGCCCTACTTT GACCCGATCCGAAATGCCGCCACCAACAATGCTCACGCAGGTGAAAGCGATGATGCATCATCCGGGTGA |
| Length | 1044 |
Transcript
| Sequence id | CopciAB_354384.T0 |
|---|---|
| Sequence |
>CopciAB_354384.T0 AAAGTGTGCGGACACCAACTCAAGTGTCCATCGACTTTCGTTTTCTCTTTGGTCTGGCGCTGGAATCCATGGGGC GCTCAACAACAAAATCGGTCGCCCGGGTGTACGCTGATGTCAACGCTCGCTTAGGGCCTTCATGGCATGAATACG ACAACCTCCAAGTTCAATGGGGATCTCAGGATCATTATGAAATCGTGCGGAAAGTGGGAAGGGGAAAGTACTCAG AGGTTTTCGAAGGAATCAATGTTGTAACGGAAGAGAAATGTATTATTAAAGTCCTCAAGCCTGTCAAGAAGAAGA AAATCAAGCGAGAAATCAAGATCCTTCAAAACTTGGCCGGAGGTCCCAATATCGTCGCTTTGCTCGATGTCGTCC GCGACCCTGCATCCAAGATCCCCAGCCTGATAACAGAATACGTGCACAACGTCGAGTTCAAGCAATTATACCCCC GATTCACCGACATGGACGTTCGCTACTACATGTTTGAACTCCTTAAGGCTCTGGACTTCTGTCACTCCAAAGGAA TCATGCACCGCGACGTTAAGCCGCACAACGTGATGATTGATCATGAGCACCGTAAATTGCGCTTGATCGACTGGG GTCTGGCTGAATTCTACCATCCAAAGACGGAATACAACGTTCGCGTGGCCTCTCGCTACTTCAAGGGACCTGAGC TCTTGGTCGACTTCCAAGAGTATGACTACAGCCTGGATATGTGGAGCTATGGTTGCATGTTCGCCTCGATGATCT TCCGCAAGGAACCCTTCTTCCACGGACACGACAACTATGACCAACTGGTTAAAATCACCAAAGTCCTTGGTACAG ACGACCTCTACGCCTATATCGAGAAGTACAACATCCGCCTTGACTCTCAATACGACGAGATTCTCGGAAGGTATC CTCGCAAGCCCTGGACCCGCTTCATCACCTCGGAAAACCAGAGATACATCTCCAACGAGGCCATCGATTTCCTTG ACAAGCTCCTCCGTTACGACCACCAAGACCGTCTCACTGCTCGTGAGGCCCAGATGCAGCCCTACTTTGACCCGA TCCGAAATGCCGCCACCAACAATGCTCACGCAGGTGAAAGCGATGATGCATCATCCGGGTGATACATACCAGCGA CACTGTATTTCTCTCTTCTTCTTCATGACTGTACGTTTGGCGATGTGTATAGAATCACTTTCTTTATTTACCCTA CCAGCATGCTTGTAAGGGCTCGGAGCAGCGTGCGGTTCTTGTTGTAATACTTTGGTATGTTGTAAAACGCCGTGT AGCTTCATTCTCATCTAATGTATACACCCTATAGGGATTCTCGTGG |
| Length | 1321 |
Gene
| Sequence id | CopciAB_354384.T0 |
|---|---|
| Sequence |
>CopciAB_354384.T0 AAAGTGTGCGGACACCAACTCAAGTGTCCATCGACTTTCGTTTTCTCTTTGGTCTGGCGCTGGAATCCATGGGGC GCTCAACAACAAAATCGGTCGCCCGGGTGTACGCTGATGTCAACGCTCGCTTAGGGCCTTCATGGCATGAATACG GTCAGTCTGCACTCCATCATTATTTGCCCTTAAACCTAACGCTTTGCTTCAGACAACCTCCAAGTTCAATGGGGA TCTCAGGATCATTATGAAATCGTGCGGAAAGTGGGAAGGGGAAAGTACTCAGAGGTTAGTTGACGTCTCTCTCAC ATAAAGGTCAGTTCTCAACATTCAATTAGGTTTTCGAAGGAATCAATGTTGTAACGGAAGAGAAATGTATTATTA AAGTCCTCAAGCCTGTCAAGAAGAAGAAAATCAAGCGAGAAATCAAGATCCTTCAAAACTTGGCCGGAGGTCCCA ATATCGTCGCTTTGCTCGATGTCGTCCGCGACCCTGCATCCAAGATCCCCAGCCTGATAACAGAATACGTGCACA ACGTCGAGTTCAAGCAATTATACCCCCGATTCACCGACATGGACGTTCGCTACTACATGTTTGAACTCCTTAAGG TATTCTCTCCATTTTTTTCTCTCCTCGAGGAGCGCTTATAGTGGCTCAGGCTCTGGACTTCTGTCACTCCAAAGG AATCATGCACCGCGACGTTAAGCCGCACAACGTGATGATTGATCATGAGCACCGTAAAGTGGGTCGTCCTTTGAC GGTTCTTTTGTAAAAAAACCGCTGAACCGATTCTCCCCGTAGTTGCGCTTGATCGACTGGGGTCTGGCTGAATTC TACCATCCAAAGACGGAATACAACGTTCGCGTGGCCTCTCGCTACTTCAAGGGACCTGAGCTCTTGGTCGACTTC CAAGAGTATGACTACAGCCTGGATATGTGGAGCTATGGTTGCATGTTCGCCTCGATGGTAACCAGTCCGGCTACC CCTTTTGTAACGTTGGCTAAATTTGCGCTCCATACAGATCTTCCGCAAGGAACCCTTCTTCCACGGACACGACAA CTATGACCAACTGGTTAAAATCACCAAAGTCCTTGGTACAGACGACCTCTACGCCTATATCGAGAAGTACAACAT CCGCCTTGACTCTCAATACGACGAGATTCTCGGAAGGTCAGAGTCTGAATTGCACCCAAAGTACGGCAGTGCAGC TGACGTGACCATGATCAGGTATCCTCGCAAGCCCTGGACCCGCTTCATCACCTCGGAAAACCAGAGATACATCTC CAACGAGGCCATCGATTTCCTTGACAAGCTCCTCCGTTACGACCACCAAGACCGTCTCACTGCTCGTGAGGCCCA GATGCAGCCCTACTTTGGTAGGTCAACTCGACTTGAGCGATCTCTCATCCGTTGACTGGATGACTGATTATTTGC CACAGACCCGATCCGAAATGCCGCCACCAACAATGCTCACGCAGGTGAAAGCGATGATGCATCATCCGGGTGATA CATACCAGCGACACTGTATTTCTCTCTTCTTCTTCATGACTGTACGTTTGGCGATGTGTATAGAATCACTTTCTT TATTTACCCTACCAGCATGCTTGTAAGGGCTCGGAGCAGCGTGCGGTTCTTGTTGTAATACTTTGGTATGTTGTA AAACGCCGTGTAGCTTCATTCTCATCTAATGTATACACCCTATAGGGATTCTCGTGG |
| Length | 1707 |