CopciAB_354501
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_354501 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 354501 |
| Uniprot id | Functional description | ||
| Location | scaffold_11:1650657..1652361 | Strand | - |
| Gene length (nt) | 1705 | Transcript length (nt) | 1568 |
| CDS length (nt) | 1335 | Protein length (aa) | 444 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| 0 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_354501.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| No records | |||||
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR032675 | Leucine-rich repeat domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
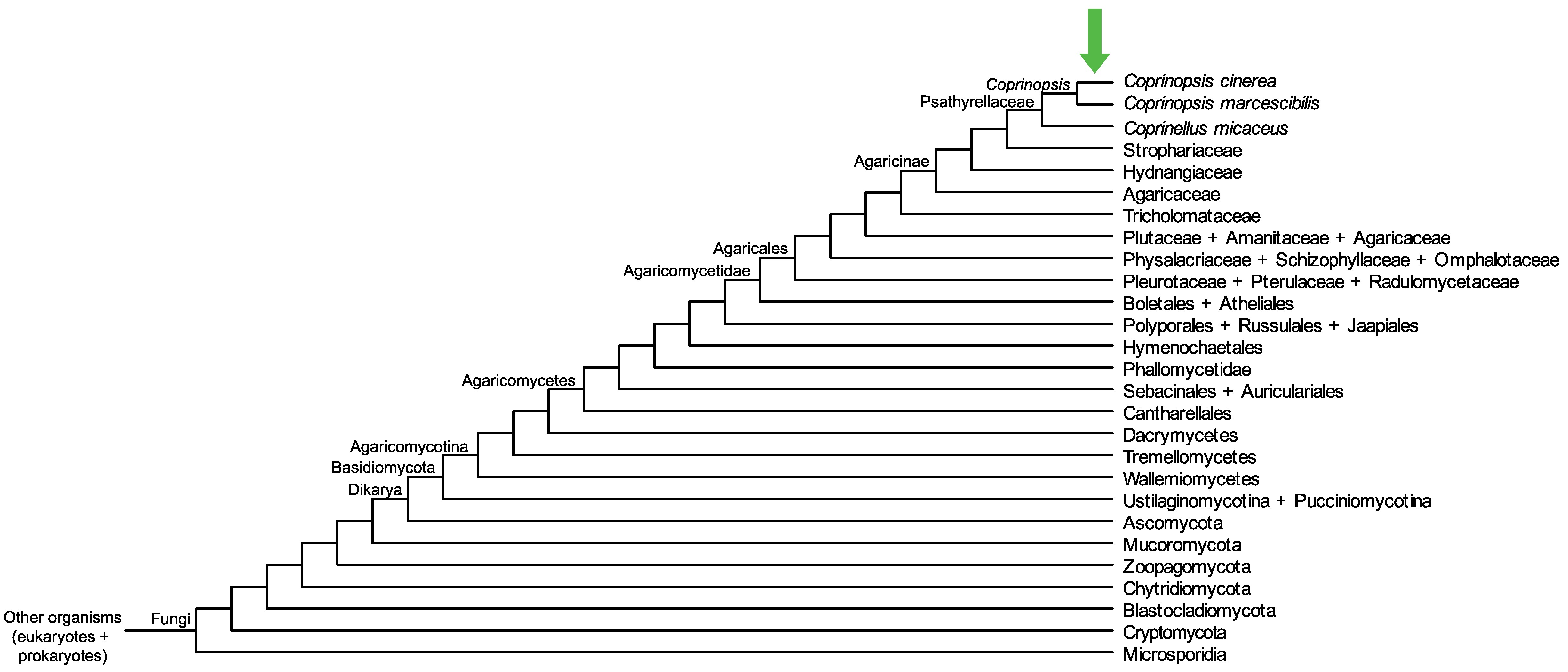
Conservation of CopciAB_354501 across fungi.
Arrow shows the origin of gene family containing CopciAB_354501.
Protein
| Sequence id | CopciAB_354501.T0 |
|---|---|
| Sequence |
>CopciAB_354501.T0 MASDPFPNELLDAVIDDLDPKTDAQTLSSLALVNQHATTIVNERRFKNVALIRSLGGRRCEGLWELVKANPNLVH NILSLQLADNSIDGGGFFFGVSDESSDEDDGDEWTSTGRVGWLGERRPVAPGLVELLTAVKPTLQVLAVCSRLQW RSLSRPVRRALLELASSPALKVLKLGRFIQIPISLFLTSPHLKQLSSKRTYISPSNSRTSNEPSQVLADELGQLD VLELRDSFSELQFLPLSLSKLRRLVVEFSPEENMFTQRMPHDVKDALWNLLEDLPLKEFRLLFVLPSHFSAQLSA LTQFILFTKWPLRVLEVAVIYRDNLDAWMGPLSLVLAKMKTLEELRLEVYPISEARSDAYDRRGATDIECRPAWE SLEKALVQLSTLRKLVIVFDERHRETEMYRDPEDTLKAAMPTLLGDGRSLRFAVEWNERPEVYYDPECE |
| Length | 444 |
Coding
| Sequence id | CopciAB_354501.T0 |
|---|---|
| Sequence |
>CopciAB_354501.T0 ATGGCCTCTGACCCCTTTCCCAACGAGCTCCTCGACGCTGTCATCGACGATCTCGACCCAAAGACAGATGCACAA ACCCTCTCCTCTCTCGCACTGGTTAACCAACACGCCACGACTATTGTCAACGAGCGCCGGTTCAAGAACGTAGCG TTAATTCGAAGCCTTGGAGGGCGTCGATGCGAAGGACTCTGGGAACTCGTCAAGGCCAACCCGAACCTCGTGCAC AACATCCTTTCGCTCCAGTTGGCCGACAATTCCATTGATGGAGGCGGTTTTTTCTTTGGAGTGTCTGATGAGTCC AGTGATGAAGACGATGGGGACGAGTGGACCTCGACTGGTAGGGTTGGATGGCTGGGAGAGAGGAGGCCTGTCGCA CCGGGTCTTGTCGAACTCTTGACGGCTGTCAAGCCCACCCTGCAGGTACTCGCAGTCTGCTCAAGACTTCAGTGG CGTAGTCTCTCGAGGCCTGTTCGACGTGCGCTCCTCGAACTCGCATCGTCGCCTGCACTCAAAGTCCTCAAACTA GGTCGCTTCATCCAAATACCTATCTCCCTCTTCCTCACCTCGCCTCACCTCAAGCAACTCTCGTCGAAAAGAACC TATATCTCCCCCTCGAATTCGCGTACCTCTAATGAGCCATCACAGGTCCTCGCCGACGAGCTAGGGCAACTCGAC GTTCTGGAGCTCAGGGACTCGTTTTCAGAGCTCCAGTTCCTCCCGTTATCTCTGTCAAAGTTGCGAAGGCTCGTC GTCGAGTTTAGCCCCGAGGAGAACATGTTCACTCAACGTATGCCCCACGACGTGAAGGACGCTTTGTGGAATTTG CTGGAAGACCTTCCGCTGAAAGAATTCCGCCTGCTTTTTGTTCTCCCGTCCCATTTCTCAGCACAGCTCTCTGCG CTCACGCAATTTATTCTCTTCACGAAATGGCCCCTTCGTGTGCTGGAGGTTGCAGTTATCTACAGAGACAATCTC GACGCTTGGATGGGGCCTTTGTCCTTGGTGCTTGCGAAAATGAAGACCCTCGAGGAGCTGCGACTCGAGGTCTAT CCGATCTCCGAAGCTCGGTCAGATGCATATGATCGAAGGGGTGCGACCGACATCGAGTGTCGCCCTGCTTGGGAG AGCCTCGAGAAAGCACTCGTCCAGCTCAGCACCCTTCGGAAGCTCGTCATCGTGTTTGACGAGAGGCATAGGGAG ACGGAGATGTATCGCGATCCTGAGGATACGTTGAAAGCGGCGATGCCAACTTTGCTTGGGGATGGTCGTTCTCTT AGGTTTGCTGTCGAATGGAACGAACGCCCCGAAGTTTATTATGACCCTGAATGTGAATAG |
| Length | 1335 |
Transcript
| Sequence id | CopciAB_354501.T0 |
|---|---|
| Sequence |
>CopciAB_354501.T0 ATCCCCAAACTGCTCGTCCAAATGGCCTCTGACCCCTTTCCCAACGAGCTCCTCGACGCTGTCATCGACGATCTC GACCCAAAGACAGATGCACAAACCCTCTCCTCTCTCGCACTGGTTAACCAACACGCCACGACTATTGTCAACGAG CGCCGGTTCAAGAACGTAGCGTTAATTCGAAGCCTTGGAGGGCGTCGATGCGAAGGACTCTGGGAACTCGTCAAG GCCAACCCGAACCTCGTGCACAACATCCTTTCGCTCCAGTTGGCCGACAATTCCATTGATGGAGGCGGTTTTTTC TTTGGAGTGTCTGATGAGTCCAGTGATGAAGACGATGGGGACGAGTGGACCTCGACTGGTAGGGTTGGATGGCTG GGAGAGAGGAGGCCTGTCGCACCGGGTCTTGTCGAACTCTTGACGGCTGTCAAGCCCACCCTGCAGGTACTCGCA GTCTGCTCAAGACTTCAGTGGCGTAGTCTCTCGAGGCCTGTTCGACGTGCGCTCCTCGAACTCGCATCGTCGCCT GCACTCAAAGTCCTCAAACTAGGTCGCTTCATCCAAATACCTATCTCCCTCTTCCTCACCTCGCCTCACCTCAAG CAACTCTCGTCGAAAAGAACCTATATCTCCCCCTCGAATTCGCGTACCTCTAATGAGCCATCACAGGTCCTCGCC GACGAGCTAGGGCAACTCGACGTTCTGGAGCTCAGGGACTCGTTTTCAGAGCTCCAGTTCCTCCCGTTATCTCTG TCAAAGTTGCGAAGGCTCGTCGTCGAGTTTAGCCCCGAGGAGAACATGTTCACTCAACGTATGCCCCACGACGTG AAGGACGCTTTGTGGAATTTGCTGGAAGACCTTCCGCTGAAAGAATTCCGCCTGCTTTTTGTTCTCCCGTCCCAT TTCTCAGCACAGCTCTCTGCGCTCACGCAATTTATTCTCTTCACGAAATGGCCCCTTCGTGTGCTGGAGGTTGCA GTTATCTACAGAGACAATCTCGACGCTTGGATGGGGCCTTTGTCCTTGGTGCTTGCGAAAATGAAGACCCTCGAG GAGCTGCGACTCGAGGTCTATCCGATCTCCGAAGCTCGGTCAGATGCATATGATCGAAGGGGTGCGACCGACATC GAGTGTCGCCCTGCTTGGGAGAGCCTCGAGAAAGCACTCGTCCAGCTCAGCACCCTTCGGAAGCTCGTCATCGTG TTTGACGAGAGGCATAGGGAGACGGAGATGTATCGCGATCCTGAGGATACGTTGAAAGCGGCGATGCCAACTTTG CTTGGGGATGGTCGTTCTCTTAGGTTTGCTGTCGAATGGAACGAACGCCCCGAAGTTTATTATGACCCTGAATGT GAATAGTATCCTCGAGCGGAGATGTAATCGCCTTTTTACCCTGCGGTAGGCTACACATAATTTTTAAATTCTCGA CCATCGCTGTCTGCATGTCGCTCTAGCCGTTGTAGTAACGAGACTCTCAGCCGTATCAGCCTTGTGTCGGAGCTC ACATTCCGCGTTTTTGCTATGTTTTACGAGCGCGTCGAGTTCGAATAAATGATAGACATTGTTCTCTC |
| Length | 1568 |
Gene
| Sequence id | CopciAB_354501.T0 |
|---|---|
| Sequence |
>CopciAB_354501.T0 ATCCCCAAACTGCTCGTCCAAATGGCCTCTGACCCCTTTCCCAACGAGCTCCTCGACGCTGTCATCGACGATCTC GACCCAAAGACAGATGCACAAACCCTCTCCTCTCTCGCACTGGTTAACCAACACGCCACGACTATTGTCAACGAG CGCCGGTTCAAGAACGTAGCGTTAATTCGAAGCCTTGGAGGGCGTCGATGCGAAGGACTCTGGGAACTCGTCAAG GCCAACCCGAACCTCGTGCACAACATCCTTTCGCTCCAGTTGGCCGACAATTCCATTGATGGAGGCGGTTTTTTC TTTGGAGTGTCTGATGAGTCCAGTGATGAAGACGATGGGGACGAGTGGACCTCGACTGGTAGGGTTGGATGGCTG GGAGAGAGGAGGCCTGTCGCACCGGGTCTTGTCGAACTCTTGACGGCTGTCAAGCCCACCCTGCAGGTACTCGCA GTCTGCTCAAGACTTCAGTGGCGTAGTCTCTCGAGGCCTGTTCGACGTGCGCTCCTCGAACTCGCATCGTCGCCT GCACTCAAAGTCCTCAAACTAGGTCGCTTCATCCAAATACCTATCTCCCTCTTCCTCACCTCGCCTCACCTCAAG CAACTCTCGTCGAAAAGAACCTATATCTCCCCCTCGAATTCGCGTACCTCTAATGAGCCATCACAGGTCCTCGCC GACGAGCTAGGGCAACTCGACGTTCTGGAGCTCAGGGACTCGTTTTCAGAGCTCCAGTTCCTCCCGTTATCTCTG TCAAAGTTGCGAAGGCTCGTCGTCGAGTTTAGCCCCGAGGAGAACATGTTCACTCAACGTATGCCCCACGACGTG AAGGACGCTTTGTGGAATTTGCTGGAAGACCTTCCGCTGAAAGAATTCCGCCTGCTTTTTGTTCTCCCGTCCCAT TTCTCAGCACAGCTCTCTGCGCTCACGCGTGAGTTTTCTCTTTCCTGATAGTTTCCCTTTTTTTGTGGAGAGCAC CGGACACTGACTTGTACGCCACCTATCTACGCAGAATTTATTCTCTTCACGAAATGGCCCCTTCGTGTGCTGGAG GTTGCAGTTATCTACAGAGACAATCTCGACGCTTGGATGGGGCCTTTGTCCTTGGTGCTTGCGAAAATGAAGACC CTCGAGGAGCTGCGACTCGAGGTCTATCCGATCTCCGAAGCTCGGTCAGATGCATATGATCGAAGGGGTGCGACC GACATCGAGTGTCGCCCTGCTTGGGAGAGCCTCGAGAAAGCACTCGTCCAGCTCAGCACCCTTCGGAAGCTCGTC ATCGTGTTTGACGAGAGGCATAGGGAGACGGAGATGTATCGCGATCCTGAGGATACGTTGAAAGCGGCGATGCCA ACTTTGCTTGGGGATGGTCGTTCTCTTAGGTTTGCTGTCGAATGGTAAGTTTTTTGAAGTCTCTTTCTGGAATGG TGGTGTTTATTCCTCTCCTTCGCAGGAACGAACGCCCCGAAGTTTATTATGACCCTGAATGTGAATAGTATCCTC GAGCGGAGATGTAATCGCCTTTTTACCCTGCGGTAGGCTACACATAATTTTTAAATTCTCGACCATCGCTGTCTG CATGTCGCTCTAGCCGTTGTAGTAACGAGACTCTCAGCCGTATCAGCCTTGTGTCGGAGCTCACATTCCGCGTTT TTGCTATGTTTTACGAGCGCGTCGAGTTCGAATAAATGATAGACATTGTTCTCTC |
| Length | 1705 |