CopciAB_356069
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_356069 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 356069 |
| Uniprot id | Functional description | atypical ABC1 ABC1-B protein kinase | |
| Location | scaffold_4:1588846..1591318 | Strand | + |
| Gene length (nt) | 2473 | Transcript length (nt) | 1995 |
| CDS length (nt) | 1884 | Protein length (aa) | 627 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Schizosaccharomyces pombe | SPBC15C4.02 |
| Aspergillus nidulans | AN4386 |
| Saccharomyces cerevisiae | YLR253W_MCP2 |
| Neurospora crassa | stk-27 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB16803 | 65.2 | 1.511E-256 | 795 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_18075 | 62.4 | 1.044E-249 | 775 |
| Lentinula edodes NBRC 111202 | Lenedo1_1108048 | 62.3 | 2.333E-249 | 774 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1065447 | 61.6 | 4.43E-249 | 773 |
| Pleurotus ostreatus PC9 | PleosPC9_1_50657 | 62.8 | 4.336E-249 | 773 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1439084 | 60.7 | 2.786E-247 | 768 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_69979 | 64.7 | 6.359E-243 | 755 |
| Grifola frondosa | Grifr_OBZ67025 | 62.3 | 1.306E-236 | 737 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_99069 | 60.4 | 8.477E-221 | 691 |
| Schizophyllum commune H4-8 | Schco3_2511118 | 55.9 | 3.838E-216 | 678 |
| Flammulina velutipes | Flave_chr07AA00789 | 62.7 | 7.503E-215 | 674 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_116507 | 63.3 | 2.706E-188 | 597 |
| Lentinula edodes B17 | Lened_B_1_1_2402 | 71.6 | 1.441E-184 | 586 |
| Auricularia subglabra | Aurde3_1_1158602 | 47.2 | 6.84E-173 | 553 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_356069.T0 |
| Description | atypical ABC1 ABC1-B protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd13969 | ADCK1-like | IPR045307 | 174 | 415 |
| Pfam | PF03109 | ABC1 atypical kinase-like domain | IPR004147 | 175 | 416 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 62 | 81 | 19 |
InterPro
| Accession | Description |
|---|---|
| IPR004147 | ABC1 atypical kinase-like domain |
| IPR045307 | ADCK1-like domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| K08869 |
EggNOG
| COG category | Description |
|---|---|
| S | atypical ABC1 ABC1-B protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
Conservation of CopciAB_356069 across fungi.
Arrow shows the origin of gene family containing CopciAB_356069.
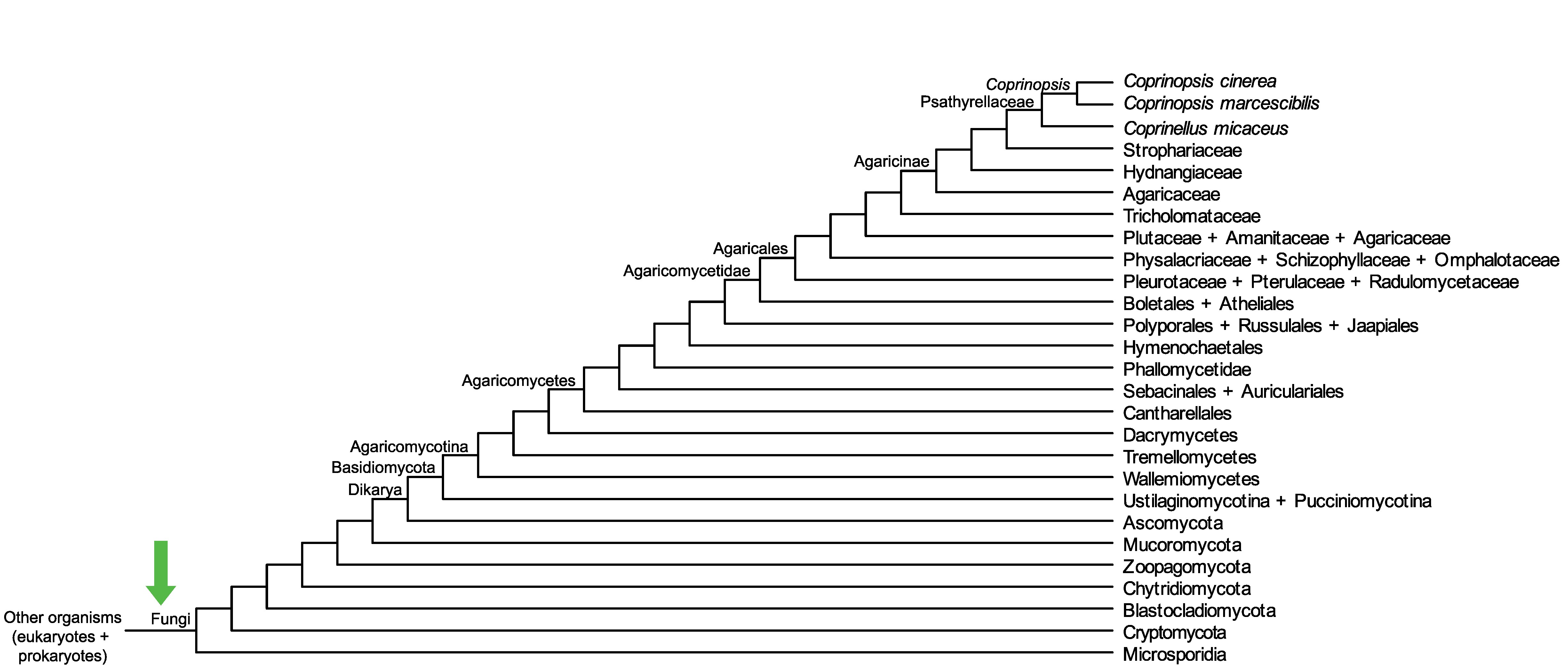
Protein
| Sequence id | CopciAB_356069.T0 |
|---|---|
| Sequence |
>CopciAB_356069.T0 MFKNGRLLQWFHSTSILRKSALSNTNTTTSRRWEQLGRRGFRSRHHTPNSSSSTATRKWRQPYIFYPACAVVLGA AGVTAYKTHKGFRHTVLAVVRCSRIADAAIRGAIDYKVMMSKSYGSDEEVNRAWSECHTRSAKRVLKALLANGGV FIKMGQHMATLVVLPVEWTSTMRPLQDQCEPTSYEELEGLFKQDMGVEVNELFEDFDPNPVGVASLAQVHVARHK PTGRRVAVKLQHPHLAEFCDIDVEMVDVTLGWIKYWFPEFEFTWLGDEMRTNLPKEMDFVHEAENAERTKRDFAH VKTSLYIPEVIHAAKRVLIMEFIEGGRVDDLEYLARHNIDRNKVAVELSRIFSEMVFHNGWFHADPHPGNLLIRP SPPSSKSPYNFEIALLDHGLYFDLDDELRVNYSRLWLSLISAASPTVIAERKRLAQLVGNVGPDLYPIFEAALTG RAALEGSFEDGKDVSFKRGSSMIDTTSRAEIEAVRHAVANTDGLILSVFDVLRRVPRRILMVLKLNDLTRGLDRA LATTHSNVRVFLVHAKYCLKASWVADQHQILSNVRTFGFTRSLCLWISGWWHYRTSYAAFAVYEALLDFQGWKVK KLAWLRGLWRQGFRGAHDAAAGLRIEA |
| Length | 627 |
Coding
| Sequence id | CopciAB_356069.T0 |
|---|---|
| Sequence |
>CopciAB_356069.T0 ATGTTCAAGAACGGTCGGCTACTTCAATGGTTCCATAGTACTTCCATTCTCCGGAAATCTGCTCTCTCGAACACT AACACGACGACTTCGCGACGTTGGGAGCAGCTTGGTCGACGTGGATTTCGATCCCGGCATCACACACCTAATTCC TCGAGTTCGACGGCGACGAGGAAATGGCGACAACCATACATCTTTTATCCCGCATGTGCGGTAGTCCTGGGGGCA GCTGGTGTCACTGCGTACAAGACGCATAAAGGGTTTAGGCACACTGTGCTGGCTGTCGTGCGGTGTTCGAGGATT GCTGACGCGGCAATACGGGGAGCGATAGATTATAAAGTTATGATGTCCAAAAGCTACGGTTCGGACGAGGAGGTG AACCGTGCATGGTCAGAATGTCATACGAGGAGTGCAAAGAGGGTTTTGAAAGCGTTGCTTGCAAACGGAGGTGTA TTCATTAAGATGGGGCAACATATGGCGACCCTCGTCGTCCTCCCTGTCGAATGGACAAGCACGATGCGGCCGTTA CAGGACCAGTGCGAACCTACATCCTACGAAGAGCTGGAAGGATTGTTTAAACAAGATATGGGAGTGGAAGTGAAC GAGCTTTTCGAGGATTTCGATCCCAATCCTGTGGGAGTTGCAAGTTTGGCCCAAGTGCATGTTGCGCGTCATAAA CCGACTGGGAGGAGAGTCGCCGTGAAGCTGCAACATCCACACTTGGCAGAGTTTTGCGATATCGATGTGGAAATG GTCGATGTTACCCTTGGGTGGATCAAGTACTGGTTCCCGGAATTCGAATTCACCTGGCTCGGGGACGAAATGAGG ACCAACCTCCCAAAAGAGATGGACTTCGTGCACGAAGCCGAGAACGCTGAACGCACAAAACGAGATTTCGCACAC GTCAAGACGTCGCTCTACATTCCTGAAGTTATCCACGCCGCGAAGCGCGTCCTGATCATGGAGTTCATCGAAGGA GGGCGAGTGGACGATTTGGAGTACCTAGCGCGTCACAATATCGACAGGAACAAGGTTGCAGTGGAACTTTCGAGG ATATTCAGCGAGATGGTGTTTCATAATGGATGGTTCCATGCTGACCCCCATCCAGGTAATTTGCTAATTCGCCCA TCGCCACCTTCGTCAAAGTCCCCGTACAACTTTGAGATCGCGTTGCTGGACCATGGACTTTACTTTGACCTTGAC GATGAACTCAGGGTGAACTATAGCAGATTGTGGCTGTCTTTGATTTCGGCAGCCTCTCCCACTGTTATCGCCGAA CGCAAGCGCCTAGCTCAACTTGTGGGCAATGTTGGTCCCGACCTTTACCCCATCTTTGAAGCGGCTCTCACTGGT CGCGCAGCTTTAGAAGGCTCGTTTGAAGATGGGAAAGATGTTTCGTTCAAGAGGGGCTCGAGCATGATCGATACA ACGAGTCGTGCTGAAATTGAGGCCGTTCGACACGCTGTCGCTAACACGGACGGGCTCATTTTGTCAGTGTTTGAT GTACTGCGAAGGGTTCCAAGGCGGATTCTTATGGTCCTGAAGTTGAATGACTTGACTCGTGGATTGGATCGTGCT CTTGCGACCACTCATTCCAATGTCCGAGTCTTCCTCGTTCACGCAAAATACTGCCTGAAGGCATCCTGGGTTGCT GACCAACACCAGATACTTTCCAATGTCCGGACTTTTGGCTTCACACGGTCGCTGTGCCTTTGGATATCTGGCTGG TGGCATTACCGTACCTCATACGCCGCCTTCGCTGTGTACGAGGCACTACTGGACTTTCAAGGTTGGAAAGTCAAG AAACTGGCATGGCTCCGGGGATTGTGGCGTCAGGGGTTTAGAGGGGCGCATGACGCTGCAGCTGGCCTGCGCATA GAAGCGTGA |
| Length | 1884 |
Transcript
| Sequence id | CopciAB_356069.T0 |
|---|---|
| Sequence |
>CopciAB_356069.T0 ACCGGACTCCACGACAGGCTCTTGAGTCCCTATGTACCATGTTCAAGAACGGTCGGCTACTTCAATGGTTCCATA GTACTTCCATTCTCCGGAAATCTGCTCTCTCGAACACTAACACGACGACTTCGCGACGTTGGGAGCAGCTTGGTC GACGTGGATTTCGATCCCGGCATCACACACCTAATTCCTCGAGTTCGACGGCGACGAGGAAATGGCGACAACCAT ACATCTTTTATCCCGCATGTGCGGTAGTCCTGGGGGCAGCTGGTGTCACTGCGTACAAGACGCATAAAGGGTTTA GGCACACTGTGCTGGCTGTCGTGCGGTGTTCGAGGATTGCTGACGCGGCAATACGGGGAGCGATAGATTATAAAG TTATGATGTCCAAAAGCTACGGTTCGGACGAGGAGGTGAACCGTGCATGGTCAGAATGTCATACGAGGAGTGCAA AGAGGGTTTTGAAAGCGTTGCTTGCAAACGGAGGTGTATTCATTAAGATGGGGCAACATATGGCGACCCTCGTCG TCCTCCCTGTCGAATGGACAAGCACGATGCGGCCGTTACAGGACCAGTGCGAACCTACATCCTACGAAGAGCTGG AAGGATTGTTTAAACAAGATATGGGAGTGGAAGTGAACGAGCTTTTCGAGGATTTCGATCCCAATCCTGTGGGAG TTGCAAGTTTGGCCCAAGTGCATGTTGCGCGTCATAAACCGACTGGGAGGAGAGTCGCCGTGAAGCTGCAACATC CACACTTGGCAGAGTTTTGCGATATCGATGTGGAAATGGTCGATGTTACCCTTGGGTGGATCAAGTACTGGTTCC CGGAATTCGAATTCACCTGGCTCGGGGACGAAATGAGGACCAACCTCCCAAAAGAGATGGACTTCGTGCACGAAG CCGAGAACGCTGAACGCACAAAACGAGATTTCGCACACGTCAAGACGTCGCTCTACATTCCTGAAGTTATCCACG CCGCGAAGCGCGTCCTGATCATGGAGTTCATCGAAGGAGGGCGAGTGGACGATTTGGAGTACCTAGCGCGTCACA ATATCGACAGGAACAAGGTTGCAGTGGAACTTTCGAGGATATTCAGCGAGATGGTGTTTCATAATGGATGGTTCC ATGCTGACCCCCATCCAGGTAATTTGCTAATTCGCCCATCGCCACCTTCGTCAAAGTCCCCGTACAACTTTGAGA TCGCGTTGCTGGACCATGGACTTTACTTTGACCTTGACGATGAACTCAGGGTGAACTATAGCAGATTGTGGCTGT CTTTGATTTCGGCAGCCTCTCCCACTGTTATCGCCGAACGCAAGCGCCTAGCTCAACTTGTGGGCAATGTTGGTC CCGACCTTTACCCCATCTTTGAAGCGGCTCTCACTGGTCGCGCAGCTTTAGAAGGCTCGTTTGAAGATGGGAAAG ATGTTTCGTTCAAGAGGGGCTCGAGCATGATCGATACAACGAGTCGTGCTGAAATTGAGGCCGTTCGACACGCTG TCGCTAACACGGACGGGCTCATTTTGTCAGTGTTTGATGTACTGCGAAGGGTTCCAAGGCGGATTCTTATGGTCC TGAAGTTGAATGACTTGACTCGTGGATTGGATCGTGCTCTTGCGACCACTCATTCCAATGTCCGAGTCTTCCTCG TTCACGCAAAATACTGCCTGAAGGCATCCTGGGTTGCTGACCAACACCAGATACTTTCCAATGTCCGGACTTTTG GCTTCACACGGTCGCTGTGCCTTTGGATATCTGGCTGGTGGCATTACCGTACCTCATACGCCGCCTTCGCTGTGT ACGAGGCACTACTGGACTTTCAAGGTTGGAAAGTCAAGAAACTGGCATGGCTCCGGGGATTGTGGCGTCAGGGGT TTAGAGGGGCGCATGACGCTGCAGCTGGCCTGCGCATAGAAGCGTGAACAATACCTTAGCGTAACAGGACCATAT CATTATATTAACGCAGTAGTAATAATACATGGATTTAGAATGACG |
| Length | 1995 |
Gene
| Sequence id | CopciAB_356069.T0 |
|---|---|
| Sequence |
>CopciAB_356069.T0 ACCGGACTCCACGACAGGCTCTTGAGTCCCTATGTACCATGTTCAAGAACGGTCGGCTACTTCAATGGTTCCATA GTACTTCCATTCTCCGGAAATCTGCTCTCTCGAACACTAACACGACGACTTCGCGACGTTGGGAGCAGCTTGGTC GACGTGGATTTCGATCCCGGCATCACACACCTAATTCCTCGAGTTCGACGGCGACGAGGAAATGGCGACAACCAT ACATCTTTTATCCCGCATGTGCGGTAGTCCTGGGGGCAGCTGGTGTCACTGCGTACAAGACGCATAAAGGGTTTA GGCACACTGTGCTGGCTGTCGTGCGGTGTTCGAGGATTGCTGGTATGGAATTTCATTCGTATTGGGAGACTGAAA TGTGGTAACGATCTTGCTCCTTAGACGCGGCAATACGGGGAGCGATAGATTATAAAGTTATGATGTCCAAAAGCT ACGGTTCGGACGAGGAGGTGAACCGTGCATGGTCAGAATGTCATACGAGGAGTGCAAAGAGGGTTTTGAAAGCGT TGCTTGCAAACGGAGGTGGGTCCCGTACTGTGGCTCAAGCTATCCGTAACCTAATCATACGATAGGTGTATTCAT TAAGATGGGGCAACATATGGCGACCCTCGTCGTCCTCCCTGTCGAATGGACAAGCACGATGCGGCCGTTACAGGA CCAGTGCGAACCTACATCCTACGAAGAGCTGGAAGGATTGTTTAAACAAGATATGGGAGTGGAAGTGAACGAGCT TTTCGAGGATTTCGATCCCAATCCTGTGGGAGTTGCAAGTTTGGCCCAAGTGCATGTTGCGCGTCATAAACCGAC TGGGAGGAGAGTCGCCGTGAAGGTGAGCAGTTGGCGATTCTTTCTGCTCGGTGAAGTTCAGGGATGCTAATTCAA AATGGACGTAGCTGCAACATCCACACTTGGCAGAGTTTTGCGATATCGATGTGGAAATGGTCGATGTTACCCTTG GTGAGTTCGTTTTGGATGAGATTATATCATTGACTCATCTACGTTACAGGGTGGATCAAGTACTGGTTCCCGGAA TTCGAATTCACCTGGCTCGGGGTAGGCATTCTTGTCCTCAGTACCTCCTCCCACTAACATACCCTAGGACGAAAT GAGGACCAACCTCCCAAAAGAGATGGACTTCGTGCACGAAGCCGAGAACGCTGAACGCACAAAACGAGATTTCGC ACACGTCAAGACGTCGCTCTACATTCCTGAAGTTATCCACGCCGCGAAGCGCGTCCTGATCATGGAGTTCATCGA AGGAGGGCGAGTGGACGATTTGGAGTACCTAGCGCGTCACAATATCGACAGGAACAAGGTTGCAGTGGAACTTTC GAGGATATTCAGCGAGATGGTGTTTCATAATGGATGGTTCCATGCTGTCAGTGCTGTCGTGTTTTGCTCATTCGA GAGGGTAATTCTGACTAGTTGAATGCAGGACCCCCATCCAGGTAATTTGCTAATTCGCCCATCGCCACCTTCGTC AAAGTCCCCGTACAACTTTGAGATCGCGTTGCTGGACCATGGACTTTACTTTGACCTTGACGATGAACTCAGGGT GAACTATAGCAGATTGTGGCTGTCTTTGATTTCGGCAGCCTCTCCCACTGTTATCGCCGAACGCAAGCGCCTAGC TCAACTTGTGGGCAATGTTGGTCCCGACCTTGTACGTCTTCACGCCCTACTGGAACACATACCCTTTCTTACCCT CACATGTAGTACCCCATCTTTGAAGCGGCTCTCACTGGTCGCGCAGCTTTAGAAGGCTCGTTTGAAGATGGGAAA GATGTTTCGTTCAAGAGGGGCTCGAGCATGATCGATACAACGAGTCGTGCTGAAATTGAGGCCGTTCGACACGCT GTCGCTAACACGGACGGGCTCATTTTGTCAGTGTTTGATGTACTGCGAAGGGTTCCAAGGCGGATTCTTATGGTC CTGAAGTTGAATGACTTGACTCGGTGAGTGGTAGCACCTCGCGATGCCCTATGGATGGGAGCTGAAAGGTTTTTG ACAGTGGATTGGATCGTGCTCTTGCGACCACTCATTCCAATGTAAGAGGACGTTTACATTCATTGACGTCATGGC TGACGAGAACAGGTCCGAGTCTTCCTCGTTCACGCAAAATACTGCCTGAAGGCATCCTGGGTTGCTGACCAACAC CAGATACTTTCCAATGTCCGGACTTTTGGCTTCACACGGTCGCTGTGCCTTTGGATATCTGGCTGGTGGCATTAC CGTACCTCATACGCCGCCTTCGCTGTGTACGAGGCACTACTGGACTTTCAAGGTTGGAAAGTCAAGAAACTGGCA TGGCTCCGGGGATTGTGGCGTCAGGGGTTTAGAGGGGCGCATGACGCTGCAGCTGGCCTGCGCATAGAAGCGTGA ACAATACCTTAGCGTAACAGGACCATATCATTATATTAACGCAGTAGTAATAATACATGGATTTAGAATGACG |
| Length | 2473 |