CopciAB_357805
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_357805 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 357805 |
| Uniprot id | Functional description | ABC1 family | |
| Location | scaffold_1:1668692..1671004 | Strand | + |
| Gene length (nt) | 2313 | Transcript length (nt) | 2151 |
| CDS length (nt) | 2022 | Protein length (aa) | 673 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YPL109C_YPL109C |
| Aspergillus nidulans | AN6996 |
| Neurospora crassa | stk-33 |
| Schizosaccharomyces pombe | SPBC21C3.03 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7265925 | 69.9 | 4.523E-292 | 929 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_124459 | 69.4 | 7.4E-292 | 928 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_182069 | 68.9 | 7.745E-301 | 925 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28402 | 67.6 | 1.017E-293 | 905 |
| Lentinula edodes NBRC 111202 | Lenedo1_1075884 | 64.3 | 1.579E-286 | 884 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_682 | 64.1 | 3.786E-285 | 880 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1063003 | 62.7 | 1.021E-280 | 867 |
| Pleurotus ostreatus PC9 | PleosPC9_1_85981 | 62.7 | 9.992E-281 | 867 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1437991 | 64 | 9.038E-275 | 850 |
| Schizophyllum commune H4-8 | Schco3_2565068 | 61.6 | 1.503E-260 | 809 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_148071 | 61.4 | 2.929E-259 | 805 |
| Flammulina velutipes | Flave_chr08AA00569 | 69.7 | 3.07E-249 | 776 |
| Auricularia subglabra | Aurde3_1_1229785 | 56.2 | 3.49E-235 | 736 |
| Grifola frondosa | Grifr_OBZ78754 | 52 | 2.187E-204 | 646 |
| Lentinula edodes B17 | Lened_B_1_1_15963 | 64.4 | 8.264E-74 | 263 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_357805.T0 |
| Description | ABC1 family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd13971 | ADCK2-like | IPR044095 | 168 | 542 |
| Pfam | PF03109 | ABC1 atypical kinase-like domain | IPR004147 | 169 | 219 |
| Pfam | PF03109 | ABC1 atypical kinase-like domain | IPR004147 | 256 | 405 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 15 | 32 | 17 |
| 2 | 93 | 112 | 19 |
InterPro
| Accession | Description |
|---|---|
| IPR004147 | ABC1 atypical kinase-like domain |
| IPR044095 | ADCK2-like dom |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| K08869 |
EggNOG
| COG category | Description |
|---|---|
| S | ABC1 family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
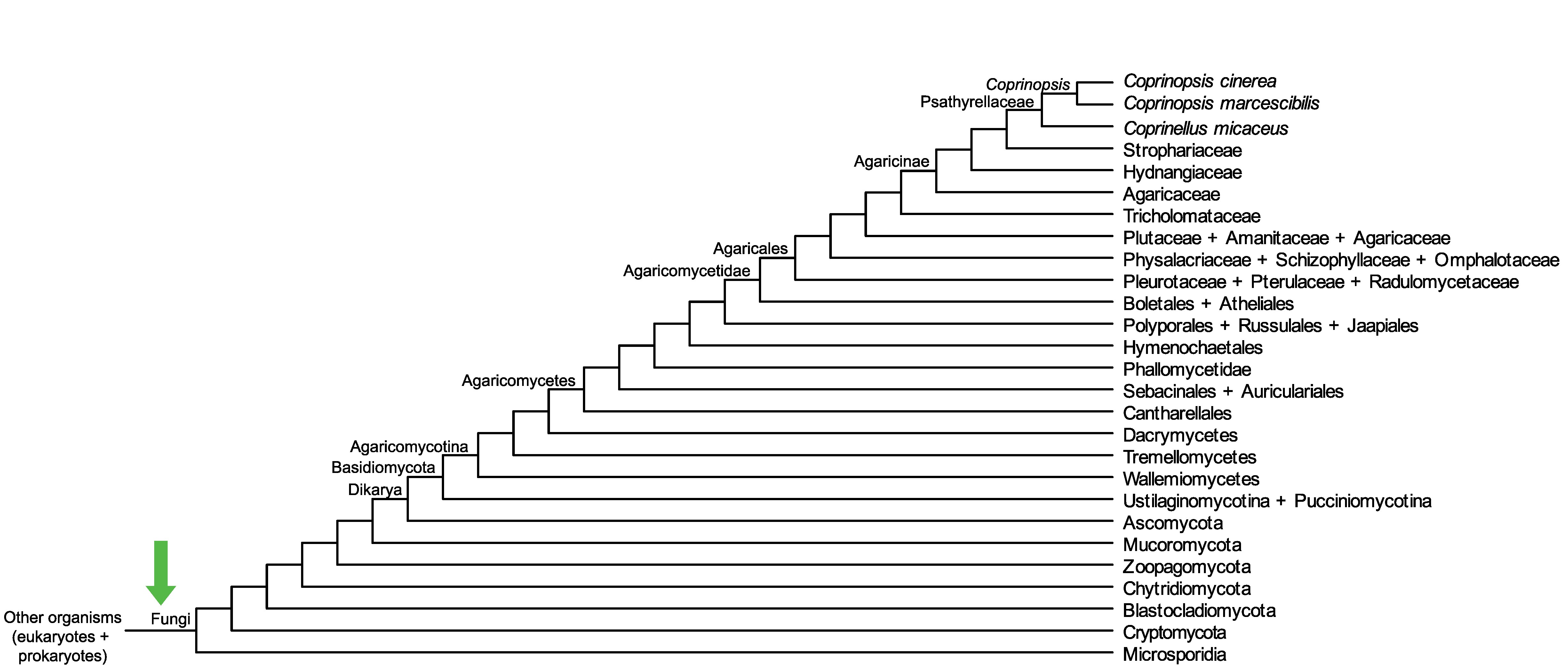
Conservation of CopciAB_357805 across fungi.
Arrow shows the origin of gene family containing CopciAB_357805.
Protein
| Sequence id | CopciAB_357805.T0 |
|---|---|
| Sequence |
>CopciAB_357805.T0 MLRAARIPRLVRSPRFLWLAAPLVVGGGVLVITRQPQTLQHSDILSSPALIPRRAALNPTILSPSEDNLTLKSSI LELLREHVWEPIRTAVRFLRLFALFFPVILSSPMLLVGRPMKEYKGDRWGAVWWYGYLVQQMEAAGPTFIKLAQW AASRVDLFPPILCERLGALHSQGTPHPFSHTKKVVEAVFQRPFDQVFEEFDQTPIGSGAIAQVYRATLKRDLIPP SYHGPRRTRQVGGALGPIILEDPPPSAPTASVAIKILHPRVEKTISRDLAIMHFFASCISLLPGMKWLSLPEEVL VFGEMMNQQLDLRHEADNLITFEANFAPRKVPVTFPRPLKIWSSKDILVEEYQNALPLETFLRNGGGPYDYQIAT VGLNAFLTMLLLDNFVHSDLHPGNIMIKFSKPPTTRILLQNLYNHFFPSSVDQSTSDVSSASLMIPADYSQSDEI VARLRKVSNSPDEWSRELLKLHSGGYVPEIVFIDAGLVTTLNDTNRTNFLDLFKAVAEFDGYKTGQLMIERSRTP ELAVDKETFALKMQHLILSIKRKTFSLGQIKIADLLTDVLKNVRKHRVKMEPDFINTVISILLLEGIGRQLDPKM DLFASALPILRQLGGQMAMDTKVEGLPSAKDLPTSDLAALLKVWVWIEARSFISGALVDMDALIQRDWLTPNI |
| Length | 673 |
Coding
| Sequence id | CopciAB_357805.T0 |
|---|---|
| Sequence |
>CopciAB_357805.T0 ATGCTCAGAGCTGCAAGAATACCAAGGCTCGTTCGTAGCCCCCGCTTCCTATGGCTTGCAGCACCCCTCGTCGTC GGTGGCGGCGTCCTCGTCATCACCAGACAGCCTCAGACCCTGCAACATAGCGACATTCTCAGCTCACCGGCTCTC ATACCGCGTCGGGCTGCACTCAACCCCACGATTCTGTCACCTTCAGAAGACAATCTTACTCTCAAGTCCAGCATT CTCGAGCTGCTCCGCGAACATGTGTGGGAGCCAATACGTACAGCAGTTCGTTTTCTGCGCCTATTCGCGCTGTTC TTTCCTGTGATCCTCTCGAGTCCAATGCTTCTCGTCGGCCGACCAATGAAAGAGTACAAAGGAGATAGATGGGGA GCTGTTTGGTGGTACGGTTACCTTGTACAGCAGATGGAAGCAGCTGGCCCGACATTCATCAAGTTGGCCCAATGG GCTGCATCCAGAGTCGACCTTTTCCCCCCTATCTTGTGCGAACGGCTCGGCGCCCTTCATTCCCAGGGAACTCCT CACCCGTTCTCTCACACAAAGAAAGTCGTCGAAGCCGTCTTCCAGCGCCCATTCGATCAGGTCTTCGAGGAATTT GATCAAACTCCAATTGGATCAGGTGCCATTGCCCAAGTTTATCGCGCAACTCTCAAGCGGGACCTCATTCCTCCT TCATATCATGGCCCCAGGCGCACCCGCCAGGTTGGTGGTGCACTTGGTCCCATCATTTTGGAGGACCCTCCCCCT TCTGCGCCGACCGCCTCGGTTGCTATCAAAATTCTTCACCCTCGAGTGGAAAAGACCATCTCCCGGGACCTAGCC ATCATGCATTTCTTCGCATCATGTATCTCCCTGCTCCCTGGCATGAAGTGGCTTTCTCTTCCAGAGGAAGTTTTG GTCTTCGGCGAGATGATGAATCAGCAGTTGGATTTGAGGCACGAAGCCGACAATCTCATTACCTTCGAAGCCAAT TTCGCCCCGAGGAAGGTTCCCGTCACCTTCCCGCGGCCATTAAAGATCTGGTCGAGTAAAGATATACTCGTCGAG GAATATCAGAACGCCTTGCCTCTGGAAACATTTTTACGAAATGGCGGTGGCCCTTACGACTATCAGATTGCGACT GTTGGTTTGAACGCGTTTTTGACCATGCTGTTACTGGATAATTTCGTCCATTCCGACCTCCACCCTGGAAATATC ATGATCAAGTTCAGCAAGCCCCCTACAACTCGAATTCTCCTCCAGAATCTGTACAACCATTTCTTCCCTAGCTCT GTCGATCAATCTACCTCCGATGTGTCGTCGGCCTCACTAATGATCCCAGCGGATTACTCCCAATCCGATGAGATT GTCGCCAGGCTACGTAAAGTCTCCAATTCGCCAGATGAATGGAGCCGGGAGCTACTCAAACTCCACTCGGGAGGA TACGTCCCCGAAATCGTCTTCATAGATGCCGGACTCGTTACCACTCTCAACGACACCAACAGAACGAACTTCCTG GACCTCTTCAAAGCTGTGGCCGAATTCGATGGATACAAGACCGGTCAGCTCATGATCGAGCGAAGCCGAACGCCT GAACTGGCCGTGGACAAGGAGACGTTTGCGTTGAAGATGCAGCACCTAATTCTCAGCATCAAGCGCAAGACGTTT AGCCTGGGACAGATTAAGATCGCCGACCTTCTAACGGACGTGTTGAAGAACGTTAGAAAGCATCGGGTGAAGATG GAGCCCGACTTTATTAACACCGTCATTTCCATTCTGTTGTTAGAAGGTATCGGGAGACAGTTGGATCCCAAGATG GACTTGTTTGCGAGTGCGTTGCCCATTCTCCGCCAACTGGGAGGGCAAATGGCGATGGATACCAAGGTGGAGGGA CTGCCTAGTGCCAAAGATCTTCCAACGAGCGATCTTGCGGCGTTGTTAAAAGTGTGGGTGTGGATAGAAGCGAGG AGTTTTATTTCTGGCGCATTGGTGGATATGGATGCGTTAATTCAGCGGGATTGGTTAACCCCGAATATTTAG |
| Length | 2022 |
Transcript
| Sequence id | CopciAB_357805.T0 |
|---|---|
| Sequence |
>CopciAB_357805.T0 AATGTTCTCGCATGCTCAGAGCTGCAAGAATACCAAGGCTCGTTCGTAGCCCCCGCTTCCTATGGCTTGCAGCAC CCCTCGTCGTCGGTGGCGGCGTCCTCGTCATCACCAGACAGCCTCAGACCCTGCAACATAGCGACATTCTCAGCT CACCGGCTCTCATACCGCGTCGGGCTGCACTCAACCCCACGATTCTGTCACCTTCAGAAGACAATCTTACTCTCA AGTCCAGCATTCTCGAGCTGCTCCGCGAACATGTGTGGGAGCCAATACGTACAGCAGTTCGTTTTCTGCGCCTAT TCGCGCTGTTCTTTCCTGTGATCCTCTCGAGTCCAATGCTTCTCGTCGGCCGACCAATGAAAGAGTACAAAGGAG ATAGATGGGGAGCTGTTTGGTGGTACGGTTACCTTGTACAGCAGATGGAAGCAGCTGGCCCGACATTCATCAAGT TGGCCCAATGGGCTGCATCCAGAGTCGACCTTTTCCCCCCTATCTTGTGCGAACGGCTCGGCGCCCTTCATTCCC AGGGAACTCCTCACCCGTTCTCTCACACAAAGAAAGTCGTCGAAGCCGTCTTCCAGCGCCCATTCGATCAGGTCT TCGAGGAATTTGATCAAACTCCAATTGGATCAGGTGCCATTGCCCAAGTTTATCGCGCAACTCTCAAGCGGGACC TCATTCCTCCTTCATATCATGGCCCCAGGCGCACCCGCCAGGTTGGTGGTGCACTTGGTCCCATCATTTTGGAGG ACCCTCCCCCTTCTGCGCCGACCGCCTCGGTTGCTATCAAAATTCTTCACCCTCGAGTGGAAAAGACCATCTCCC GGGACCTAGCCATCATGCATTTCTTCGCATCATGTATCTCCCTGCTCCCTGGCATGAAGTGGCTTTCTCTTCCAG AGGAAGTTTTGGTCTTCGGCGAGATGATGAATCAGCAGTTGGATTTGAGGCACGAAGCCGACAATCTCATTACCT TCGAAGCCAATTTCGCCCCGAGGAAGGTTCCCGTCACCTTCCCGCGGCCATTAAAGATCTGGTCGAGTAAAGATA TACTCGTCGAGGAATATCAGAACGCCTTGCCTCTGGAAACATTTTTACGAAATGGCGGTGGCCCTTACGACTATC AGATTGCGACTGTTGGTTTGAACGCGTTTTTGACCATGCTGTTACTGGATAATTTCGTCCATTCCGACCTCCACC CTGGAAATATCATGATCAAGTTCAGCAAGCCCCCTACAACTCGAATTCTCCTCCAGAATCTGTACAACCATTTCT TCCCTAGCTCTGTCGATCAATCTACCTCCGATGTGTCGTCGGCCTCACTAATGATCCCAGCGGATTACTCCCAAT CCGATGAGATTGTCGCCAGGCTACGTAAAGTCTCCAATTCGCCAGATGAATGGAGCCGGGAGCTACTCAAACTCC ACTCGGGAGGATACGTCCCCGAAATCGTCTTCATAGATGCCGGACTCGTTACCACTCTCAACGACACCAACAGAA CGAACTTCCTGGACCTCTTCAAAGCTGTGGCCGAATTCGATGGATACAAGACCGGTCAGCTCATGATCGAGCGAA GCCGAACGCCTGAACTGGCCGTGGACAAGGAGACGTTTGCGTTGAAGATGCAGCACCTAATTCTCAGCATCAAGC GCAAGACGTTTAGCCTGGGACAGATTAAGATCGCCGACCTTCTAACGGACGTGTTGAAGAACGTTAGAAAGCATC GGGTGAAGATGGAGCCCGACTTTATTAACACCGTCATTTCCATTCTGTTGTTAGAAGGTATCGGGAGACAGTTGG ATCCCAAGATGGACTTGTTTGCGAGTGCGTTGCCCATTCTCCGCCAACTGGGAGGGCAAATGGCGATGGATACCA AGGTGGAGGGACTGCCTAGTGCCAAAGATCTTCCAACGAGCGATCTTGCGGCGTTGTTAAAAGTGTGGGTGTGGA TAGAAGCGAGGAGTTTTATTTCTGGCGCATTGGTGGATATGGATGCGTTAATTCAGCGGGATTGGTTAACCCCGA ATATTTAGAACAGGTTTACATGTTCATCACAAAGACAGTAGTCACAACCAGTGGCTTTCGGTTTCACTCCGCTGC TCTCACCATCTGTATTGTATAGCCTATCTATAGACTCTCAACTTAAGTCCC |
| Length | 2151 |
Gene
| Sequence id | CopciAB_357805.T0 |
|---|---|
| Sequence |
>CopciAB_357805.T0 AATGTTCTCGCATGCTCAGAGCTGCAAGAATACCAAGGCTCGTTCGTAGCCCCCGCTTCCTATGGCTTGCAGCAC CCCTCGTCGTCGGTGGCGGCGTCCTCGTCATCACCAGACAGCCTCAGACCCTGCAACATAGCGACATTCTCAGCT CACCGGCTCTCATACCGCGTCGGGCTGCACTCAACCCCACGATTCTGTCACCTTCAGAAGACAATCTTACTCTCA AGTCCAGCATTCTCGAGCTGCTCCGCGAACATGTGTGGGAGCCAATACGTACAGCAGTTCGTTTTCTGCGCCTAT TCGCGCTGTTCTTTCCTGTGATCCTCTCGAGTCCAATGCTTCTCGTCGGCCGACCAATGAAAGAGTACAAAGGAG ATAGATGGGGAGCTGTTTGGTGGTACGGTTACCTTGTACAGCAGATGGAAGCAGCTGGCCCGACATTCATCAAGG TATGTGTTCTCTCCCTGATCCCAGCTCAAATAAGCTCATCCTTCTTCCAATTCAGTTGGCCCAATGGGCTGCATC CAGAGTCGACCTTTTCCCCCCTATCTTGTGCGAACGGCTCGGCGCCCTTCATTCCCAGGGAACTCCTCACCCGTT CTCTCACACAAAGAAAGTCGTCGAAGCCGTCTTCCAGCGCCCATTCGATCAGGTCTTCGAGGAATTTGATCAAAC TCCAATTGGATCAGGTGCCATTGCCCAAGTTTATCGCGCAACTCTCAAGCGGGACCTCATTCCTCCTTCATATCA TGGCCCCAGGCGCACCCGCCAGGTTGGTGGTGCACTTGGTCCCATCATTTTGGAGGACCCTCCCCCTTCTGCGCC GACCGCCTCGGTTGCTATCAAAATTCTTCACCCTCGAGTGGAAAAGACCATCTCCCGGGACCTAGCCATCATGCA TTTCTTCGCATCATGTATCTCCCTGCTCCCTGGCATGAAGTGGCTTTCTCTTCCAGAGGAAGTTTTGGTCTTCGG CGAGATGATGAATCAGCAGTTGGATTTGAGGCACGAAGCCGACAATCTCATTACCTTCGAAGCCAATTTCGCCCC GAGGAAGGTTCCCGTCACCTTCCCGCGGCCATTAAAGATCTGGTCGAGTAAAGATATACTCGTCGAGGAATATCA GAACGCCTTGCCTCTGGAAACATTTTTACGAAATGGCGGTGGCCCTTACGACTATCAGATTGCGACTGTTGGTTT GAACGCGTTTTTGGTGCGCATTTGCCCCCATGAAATTCCCTTCCCTTTCTGATCCATCTCTAGACCATGCTGTTA CTGGATAATTTCGTCCATTCCGACCTCCACCCTGGAAATATCATGATCAAGTTCAGCAAGCCCCCTACAACTCGA ATTCTCCTCCAGAATCTGTACAACCATTTCTTCCCTAGCTCTGTCGATCAATCTACCTCCGATGTGTCGTCGGCC TCACTAATGATCCCAGCGGATTACTCCCAATCCGATGAGATTGTCGCCAGGCTACGTAAAGTCTCCAATTCGCCA GATGAATGGAGCCGGGAGCTACTCAAACTCCACTCGGGAGGATACGTCCCCGAAATCGTCTTCATAGATGCCGGA CTCGTTACCACTCTCAACGACACCAACAGAACGAACTTCCTGGACCTCTTCAAAGCTGTGGCCGAATTCGATGGA TACAAGACCGGTCAGCTCATGATCGAGCGAAGCCGAACGCCTGAACTGGCCGTGGACAAGGAGACGTTTGCGTTG AAGATGCAGCACCTAATTCTCAGCATCAAGCGCAAGACGTTTAGCCTGGGACAGATTAAGATCGCCGACCTTCTA ACGGACGTGTTGAAGAACGTTAGAAAGCATCGGGTGAAGATGGAGCCCGACTTTATTAACACCGTCATTTCCATT CTGTTGTTAGAAGGTATCGGGAGACAGTTGGATCCCAAGATGGACTTGTTTGCGAGTGCGTTGCCCATTCTCCGC CAACTGGGAGGGCAAATGGCGATGGATACCAAGGTGGAGGGACTGCCTAGTGCCAAAGATCTTCCAACGAGCGAT CTTGCGGCGTTGTTAAAAGTGTGGGTGTGGATAGAAGCGAGGAGTTTTATTTCTGGCGCATTGGTGGATATGGAT GCGTTAATTCAGCGGGATTGGTGGGTCCTCTCTTCTGTGAGGACGGTCCCGACGGATGACTGACATGGCTTTACA GGTTAACCCCGAATATTTAGAACAGGTTTACATGTTCATCACAAAGACAGTAGTCACAACCAGTGGCTTTCGGTT TCACTCCGCTGCTCTCACCATCTGTATTGTATAGCCTATCTATAGACTCTCAACTTAAGTCCC |
| Length | 2313 |