CopciAB_361063
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_361063 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | PKA4 | Synonyms | 361063 |
| Uniprot id | Functional description | Extension to Ser/Thr-type protein kinases | |
| Location | scaffold_2:2182849..2185286 | Strand | + |
| Gene length (nt) | 2438 | Transcript length (nt) | 2275 |
| CDS length (nt) | 1374 | Protein length (aa) | 457 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7267935 | 73.2 | 2.004E-221 | 684 |
| Lentinula edodes B17 | Lened_B_1_1_11778 | 68.4 | 1.274E-205 | 638 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11004 | 68.4 | 1.598E-205 | 638 |
| Flammulina velutipes | Flave_chr09AA00133 | 72.2 | 7.319E-205 | 636 |
| Lentinula edodes NBRC 111202 | Lenedo1_1039442 | 83.7 | 4.263E-203 | 631 |
| Pleurotus ostreatus PC9 | PleosPC9_1_114122 | 66.3 | 4.655E-200 | 622 |
| Schizophyllum commune H4-8 | Schco3_2484274 | 71.1 | 6.878E-199 | 619 |
| Pleurotus ostreatus PC15 | PleosPC15_2_19948 | 80.8 | 7.375E-195 | 607 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_154065 | 64.1 | 1.752E-194 | 606 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1380818 | 80.8 | 1.924E-194 | 606 |
| Grifola frondosa | Grifr_OBZ75731 | 80.7 | 1.333E-188 | 589 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_64256 | 72.4 | 1.258E-177 | 557 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_34901 | 72.4 | 1.31E-177 | 557 |
| Auricularia subglabra | Aurde3_1_1325074 | 70.2 | 6.804E-157 | 498 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | PKA4 |
|---|---|
| Protein id | CopciAB_361063.T0 |
| Description | Extension to Ser/Thr-type protein kinases |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd05580 | STKc_PKA_like | - | 131 | 423 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 134 | 393 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
| IPR000961 | AGC-kinase, C-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K04345 |
| K08282 |
| K19584 |
EggNOG
| COG category | Description |
|---|---|
| T | Extension to Ser/Thr-type protein kinases |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
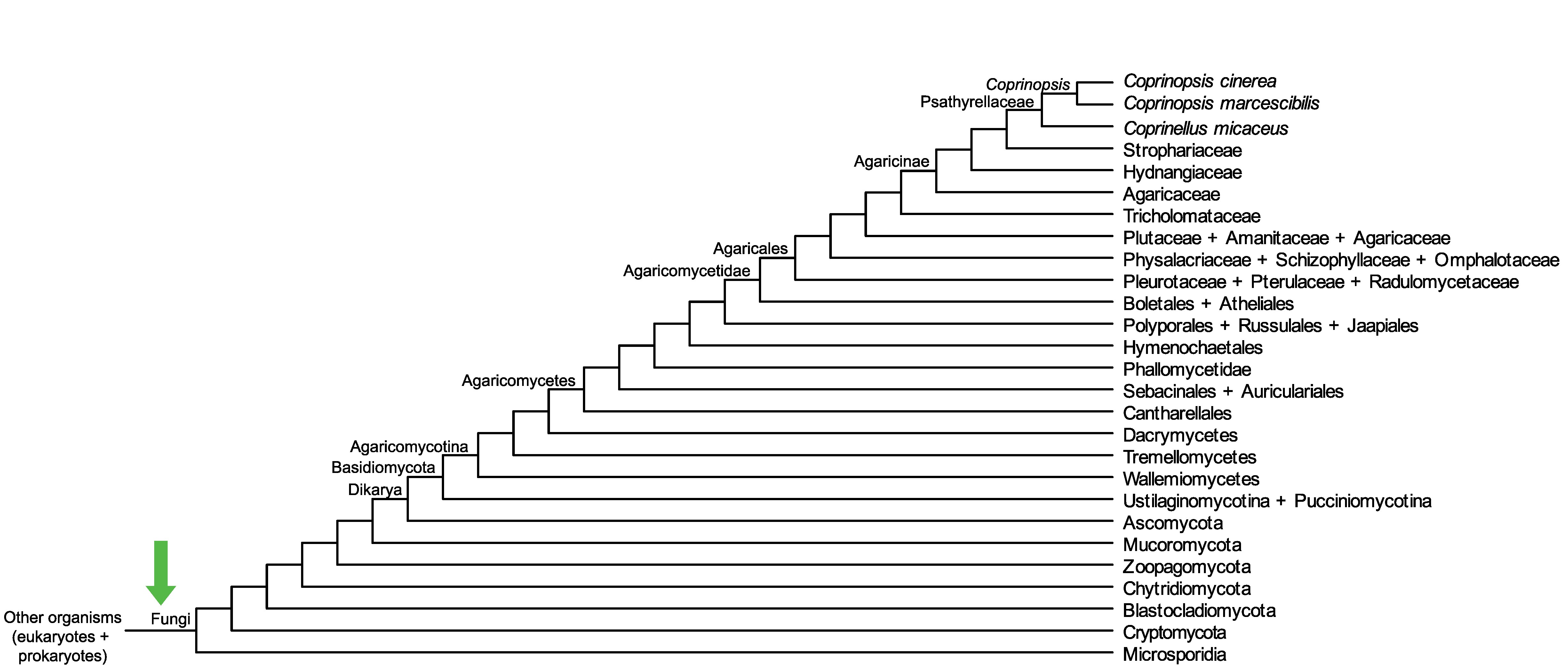
Conservation of CopciAB_361063 across fungi.
Arrow shows the origin of gene family containing CopciAB_361063.
Protein
| Sequence id | CopciAB_361063.T0 |
|---|---|
| Sequence |
>CopciAB_361063.T0 MSFQHHPTSLSMPAEPVRRISITYQQPSCPESQVVVTEAPITPPLSPKSIATKSGAMPVEEMPICSSKPIQIHSE QEAMDVESAPVHQHPNTMPGEQQAQSQDQSNESQNRPQRCLEDQRVRLQGSGLKLSDFEVRGTLGTGTFGKVLLV RHKTARDRHGAQSHFAMKVLRKTVIVQLRQVEHVNAERYILSRVNHPFIVDLFATFQDSLNVYMLMSYVPGGELF THLRRAQRFTPDVTRFYLATIILALKYLHSFNIIYRDLKPENLLLDSRGYLRLTDFGFAKIVDDRTWTLCGTPEY LAPEIIQSDGHGKAADWWACGVLCYEMLVGYPPFFDESPYGIYEKILNGQIHWPRHIDRLSKDLIKSFLNPDRTK RLGNMLGGPQDILDHPWFRGVDWDALERREINAPIIPRTTSVDDTRHFLHLPPPTAEEIPGLVGQDLPQPRFDPI NYHFLEF |
| Length | 457 |
Coding
| Sequence id | CopciAB_361063.T0 |
|---|---|
| Sequence |
>CopciAB_361063.T0 ATGTCCTTCCAGCATCACCCAACATCGTTATCAATGCCCGCTGAACCCGTTCGGCGGATATCGATTACATACCAG CAGCCTTCGTGTCCAGAATCGCAGGTCGTAGTCACTGAGGCCCCCATAACTCCTCCCCTTTCTCCCAAGTCCATC GCCACAAAGTCGGGCGCTATGCCCGTGGAAGAAATGCCCATCTGCAGTTCGAAACCTATTCAAATACACTCCGAA CAGGAGGCGATGGACGTCGAGTCGGCGCCCGTTCACCAACATCCGAATACCATGCCAGGCGAACAACAAGCCCAG TCCCAAGATCAGTCTAACGAATCGCAGAACCGCCCACAACGGTGTCTTGAAGACCAGCGTGTTCGGTTGCAGGGT TCTGGTTTGAAGCTGTCCGATTTTGAGGTCAGGGGAACACTTGGCACCGGGACGTTTGGGAAGGTACTCTTGGTG CGCCATAAGACCGCTCGAGACCGACATGGAGCGCAAAGTCATTTTGCGATGAAAGTGCTTCGCAAAACGGTGATC GTGCAACTGCGACAAGTCGAGCACGTCAATGCGGAACGATATATCCTCTCGAGGGTGAACCATCCATTCATCGTG GACCTCTTTGCGACCTTCCAGGACAGTCTCAACGTCTATATGCTCATGTCCTATGTCCCTGGGGGTGAACTTTTC ACCCATCTGCGACGCGCACAGCGGTTCACTCCTGACGTTACCCGATTCTACCTTGCCACGATTATCCTCGCCCTC AAGTACCTCCACTCGTTCAACATCATCTACCGCGACTTGAAGCCCGAAAACCTTCTGTTGGATTCTCGGGGTTAT CTCCGACTCACAGACTTTGGCTTCGCGAAAATTGTCGACGACCGAACTTGGACTCTGTGTGGCACCCCAGAATAC CTTGCCCCAGAGATTATCCAATCCGACGGTCACGGCAAGGCCGCGGATTGGTGGGCATGTGGGGTGTTGTGCTAC GAGATGCTGGTTGGATATCCGCCCTTCTTCGACGAATCTCCGTACGGCATTTACGAAAAGATTCTGAACGGCCAA ATCCACTGGCCGCGGCACATTGATCGTCTTTCAAAAGACTTGATCAAGTCGTTCCTCAACCCAGATCGCACCAAG CGACTGGGTAACATGCTGGGAGGCCCCCAGGATATCCTTGACCATCCCTGGTTTAGAGGTGTTGATTGGGATGCA TTGGAAAGGCGTGAAATCAACGCTCCCATTATCCCAAGGACGACATCGGTCGACGATACCCGCCACTTCTTGCAT CTCCCTCCTCCTACCGCTGAGGAAATCCCCGGCCTTGTTGGGCAAGATCTCCCTCAGCCACGCTTTGACCCGATC AATTACCACTTCCTCGAGTTTTAA |
| Length | 1374 |
Transcript
| Sequence id | CopciAB_361063.T0 |
|---|---|
| Sequence |
>CopciAB_361063.T0 GCCCTTTGCTTCCTCACCACCACCACCTCACTCCAAATCCGCCCCCTACACTTCGCCGTCTTCAACGACGACGTC CTCTTTCCCTGCCATTTCGCAGGCTTTTCTCGCTTCTCCTCAATAAATTCTCCACCTTCCTTTATTTTATTTCTC TTCTTTTCTCTTCTTCTCCTCACTCGTTGCTCGAGCTGCGGTAGGAGCCCAACATCTCAGCCCCCACCCCACCCA ACATAAATACCAGTCGCCATTAGAACCTTTAGCGGCCGTTGCGCACATCTTCCCTTATTCCCGGTCCCAAATGTC CTTCCAGCATCACCCAACATCGTTATCAATGCCCGCTGAACCCGTTCGGCGGATATCGATTACATACCAGCAGCC TTCGTGTCCAGAATCGCAGGTCGTAGTCACTGAGGCCCCCATAACTCCTCCCCTTTCTCCCAAGTCCATCGCCAC AAAGTCGGGCGCTATGCCCGTGGAAGAAATGCCCATCTGCAGTTCGAAACCTATTCAAATACACTCCGAACAGGA GGCGATGGACGTCGAGTCGGCGCCCGTTCACCAACATCCGAATACCATGCCAGGCGAACAACAAGCCCAGTCCCA AGATCAGTCTAACGAATCGCAGAACCGCCCACAACGGTGTCTTGAAGACCAGCGTGTTCGGTTGCAGGGTTCTGG TTTGAAGCTGTCCGATTTTGAGGTCAGGGGAACACTTGGCACCGGGACGTTTGGGAAGGTACTCTTGGTGCGCCA TAAGACCGCTCGAGACCGACATGGAGCGCAAAGTCATTTTGCGATGAAAGTGCTTCGCAAAACGGTGATCGTGCA ACTGCGACAAGTCGAGCACGTCAATGCGGAACGATATATCCTCTCGAGGGTGAACCATCCATTCATCGTGGACCT CTTTGCGACCTTCCAGGACAGTCTCAACGTCTATATGCTCATGTCCTATGTCCCTGGGGGTGAACTTTTCACCCA TCTGCGACGCGCACAGCGGTTCACTCCTGACGTTACCCGATTCTACCTTGCCACGATTATCCTCGCCCTCAAGTA CCTCCACTCGTTCAACATCATCTACCGCGACTTGAAGCCCGAAAACCTTCTGTTGGATTCTCGGGGTTATCTCCG ACTCACAGACTTTGGCTTCGCGAAAATTGTCGACGACCGAACTTGGACTCTGTGTGGCACCCCAGAATACCTTGC CCCAGAGATTATCCAATCCGACGGTCACGGCAAGGCCGCGGATTGGTGGGCATGTGGGGTGTTGTGCTACGAGAT GCTGGTTGGATATCCGCCCTTCTTCGACGAATCTCCGTACGGCATTTACGAAAAGATTCTGAACGGCCAAATCCA CTGGCCGCGGCACATTGATCGTCTTTCAAAAGACTTGATCAAGTCGTTCCTCAACCCAGATCGCACCAAGCGACT GGGTAACATGCTGGGAGGCCCCCAGGATATCCTTGACCATCCCTGGTTTAGAGGTGTTGATTGGGATGCATTGGA AAGGCGTGAAATCAACGCTCCCATTATCCCAAGGACGACATCGGTCGACGATACCCGCCACTTCTTGCATCTCCC TCCTCCTACCGCTGAGGAAATCCCCGGCCTTGTTGGGCAAGATCTCCCTCAGCCACGCTTTGACCCGATCAATTA CCACTTCCTCGAGTTTTAAGTTCTCGGGTCACCCTTTTCCTTGCTCTTTTCGACCATCTCTGAATGGATCCCACG CTCTTTCGCCTTTCCCAGTCTCCACTCTTTAATCCATCTTCTGAATCACTTTATTACGCGCCTCGGTTCAGCCTT ATCCCTATTTATTGCATTCGCATCTTCGTTCGTATCTTACCCTGCAGCATCTGGAATCCCCTATTTCTGTCTCGA AAACTGTATCCAATTCTGCCTCTCTCTTTGGTGGCTATTCTTATCTTGGTGCCACATCCCACTGTATTTTTCGCC TCCCAATCTTGCGTATGCTCTCTGTATTGCCCATTCCATTCCTTTTGTGATTTGCGGCAGTAGAACATAGTTGTT TCTCTCTGCTTTCGATGTCCTGTAGCCTTTCACTTTTTCTTGCATGTTGTTCTCTACAACACCCACCCTATGTAG CCTTGTCATCTACTCGTCACATCCTCTGGATGTTTGACTGCTGTGTATTTTTGCCATCCATATACCACTTTCGAG TCTCGTATTTCCGTCGCTGTTAGGTGTGCGACGTAATCATATATCCCCTATATTACTTGATACTTGACCTTCGCT TTTATCAATGCTACTATTTGGCTTG |
| Length | 2275 |
Gene
| Sequence id | CopciAB_361063.T0 |
|---|---|
| Sequence |
>CopciAB_361063.T0 GCCCTTTGCTTCCTCACCACCACCACCTCACTCCAAATCCGCCCCCTACACTTCGCCGTCTTCAACGACGACGTC CTCTTTCCCTGCCATTTCGCAGGCTTTTCTGTAAGTCATTTGCGCATCGCGACCACGAGCGCTTTGTTGATCAAA CAATAGCGCTTCTCCTCAATAAATTCTCCACCTTCCTTTATTTTATTTCTCTTCTTTTCTCTTCTTCTCCTCACT CGTTGCTCGAGCTGCGGTAGGAGCCCAACATCTCAGCCCCCACCCCACCCAACATAAATACCAGTCGCCATTAGA ACCTTTAGCGGCCGTTGCGCACATCTTCCCTTATTCCCGGTCCCAAATGTCCTTCCAGCATCACCCAACATCGTT ATCAATGCCCGCTGAACCCGTTCGGCGGATATCGATTACATACCAGCAGCCTTCGTGTCCAGAATCGCAGGTCGT AGTCACTGAGGCCCCCATAACTCCTCCCCTTTCTCCCAAGTCCATCGCCACAAAGTCGGGCGCTATGCCCGTGGA AGAAATGCCCATCTGCAGTTCGAAACCTATTCAAATACACTCCGAACAGGAGGCGATGGACGTCGAGTCGGCGCC CGTTCACCAACATCCGAATACCATGCCAGGCGAACAACAAGCCCAGTCCCAAGATCAGTCTAACGAATCGCAGAA CCGCCCACAACGGTGTCTTGAAGACCAGCGTGTTCGGTTGCAGGGTTCTGGTTTGAAGCTGTCCGATTTTGAGGT CAGGGGAACACTTGGTAAGTTCATTTTTGGTTTTCGTCCCATAATTCCCAGCTCACAAGAGGGTACTTCATATAG GCACCGGGACGTTTGGGAAGGTACTCTTGGTGCGCCATAAGACCGCTCGAGACCGACATGGAGCGCAAAGTCATT TTGCGATGAAAGTGCTTCGCAAAACGGTGATCGTGCAACTGCGACAAGTCGAGCACGTCAATGCGGAACGATATA TCCTCTCGAGGGTGAACCATCCATTCATCGTGGACCTCTTTGCGACCTTCCAGGACAGTCTCAACGTCTATATGC TCATGTCCTATGTCCCTGGGGGTGAACTTTTCACCCATCTGCGACGCGCACAGCGGTTCACTCCTGACGTTACCC GATTCTACCTTGCCACGATTATCCTCGCCCTCAAGTACCTCCACTCGTTCAACATCATCTACCGCGACTTGAAGC CCGAAAACCTTCTGTTGGATTCTCGGGGTTATCTCCGACTCACAGACTTTGGCTTCGCGAAAATTGTCGACGACC GAACTTGGACTCTGTGTGGCACCCCAGAATACCTTGCCCCAGAGATTATCCAATCCGACGGTCACGGCAAGGCCG CGGATTGGTGGGCATGTGGGGTGTTGTGCTACGAGATGCTGGTTGGATATCCGCCCTTCTTCGACGAATCTCCGT ACGGCATTTACGAAAAGATTCTGAACGGCCAAATCCACTGGCCGCGGCACATTGATCGTCTTTCAAAAGACTTGA TCAAGTCGTTCCTCAACCCAGATCGCACCAAGCGACTGGGTAACATGCTGGGAGGCCCCCAGGATATCCTTGACC ATCCCTGGTTTAGAGGTGTTGATTGGGATGCATTGGAAAGGCGTGAAATCAACGTATGTAGTGTTTACTCGTCTC CCTCTTCTTTTCGCTTAACCGTTGCGTAGGCTCCCATTATCCCAAGGACGACATCGGTCGACGATACCCGCCACT TCTTGCATCTCCCTCCTCCTACCGCTGAGGAAATCCCCGGCCTTGTTGGGCAAGATCTCCCTCAGCCACGCTTTG ACCCGATCAATTACCACTTCCTCGAGTTTTAAGTTCTCGGGTCACCCTTTTCCTTGCTCTTTTCGACCATCTCTG AATGGATCCCACGCTCTTTCGCCTTTCCCAGTCTCCACTCTTTAATCCATCTTCTGAATCACTTTATTACGCGCC TCGGTTCAGCCTTATCCCTATTTATTGCATTCGCATCTTCGTTCGTATCTTACCCTGCAGCATCTGGAATCCCCT ATTTCTGTCTCGAAAACTGTATCCAATTCTGCCTCTCTCTTTGGTGGCTATTCTTATCTTGGTGCCACATCCCAC TGTATTTTTCGCCTCCCAATCTTGCGTATGCTCTCTGTATTGCCCATTCCATTCCTTTTGTGATTTGCGGCAGTA GAACATAGTTGTTTCTCTCTGCTTTCGATGTCCTGTAGCCTTTCACTTTTTCTTGCATGTTGTTCTCTACAACAC CCACCCTATGTAGCCTTGTCATCTACTCGTCACATCCTCTGGATGTTTGACTGCTGTGTATTTTTGCCATCCATA TACCACTTTCGAGTCTCGTATTTCCGTCGCTGTTAGGTGTGCGACGTAATCATATATCCCCTATATTACTTGATA CTTGACCTTCGCTTTTATCAATGCTACTATTTGGCTTG |
| Length | 2438 |