CopciAB_361663
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_361663 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | KIC1 | Synonyms | 361663 |
| Uniprot id | Functional description | Ste ste20 ysk protein kinase | |
| Location | scaffold_4:2312569..2314959 | Strand | - |
| Gene length (nt) | 2391 | Transcript length (nt) | 2243 |
| CDS length (nt) | 2199 | Protein length (aa) | 732 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Lentinula edodes B17 | Lened_B_1_1_11774 | 45 | 1.528E-206 | 655 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11000 | 45 | 1.916E-206 | 655 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB16854 | 45.4 | 6.765E-202 | 642 |
| Flammulina velutipes | Flave_chr08AA01194 | 46.1 | 2.619E-190 | 608 |
| Agrocybe aegerita | Agrae_CAA7259300 | 73.6 | 1.745E-175 | 565 |
| Lentinula edodes NBRC 111202 | Lenedo1_1159371 | 84.8 | 2.855E-167 | 541 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_115914 | 40.1 | 1.357E-165 | 536 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1379378 | 82.7 | 7.705E-164 | 531 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_70261 | 80.5 | 1.416E-161 | 524 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_36853 | 80.5 | 1.476E-161 | 524 |
| Grifola frondosa | Grifr_OBZ68148 | 66 | 1.397E-152 | 498 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1065193 | 83.7 | 3.078E-137 | 453 |
| Pleurotus ostreatus PC9 | PleosPC9_1_112209 | 84.6 | 3.013E-137 | 453 |
| Schizophyllum commune H4-8 | Schco3_2738395 | 64.6 | 6.136E-129 | 429 |
| Auricularia subglabra | Aurde3_1_72597 | 64.3 | 1.158E-122 | 411 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | KIC1 |
|---|---|
| Protein id | CopciAB_361663.T0 |
| Description | Ste ste20 ysk protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 10 | 266 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08286 |
EggNOG
| COG category | Description |
|---|---|
| T | Ste ste20 ysk protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
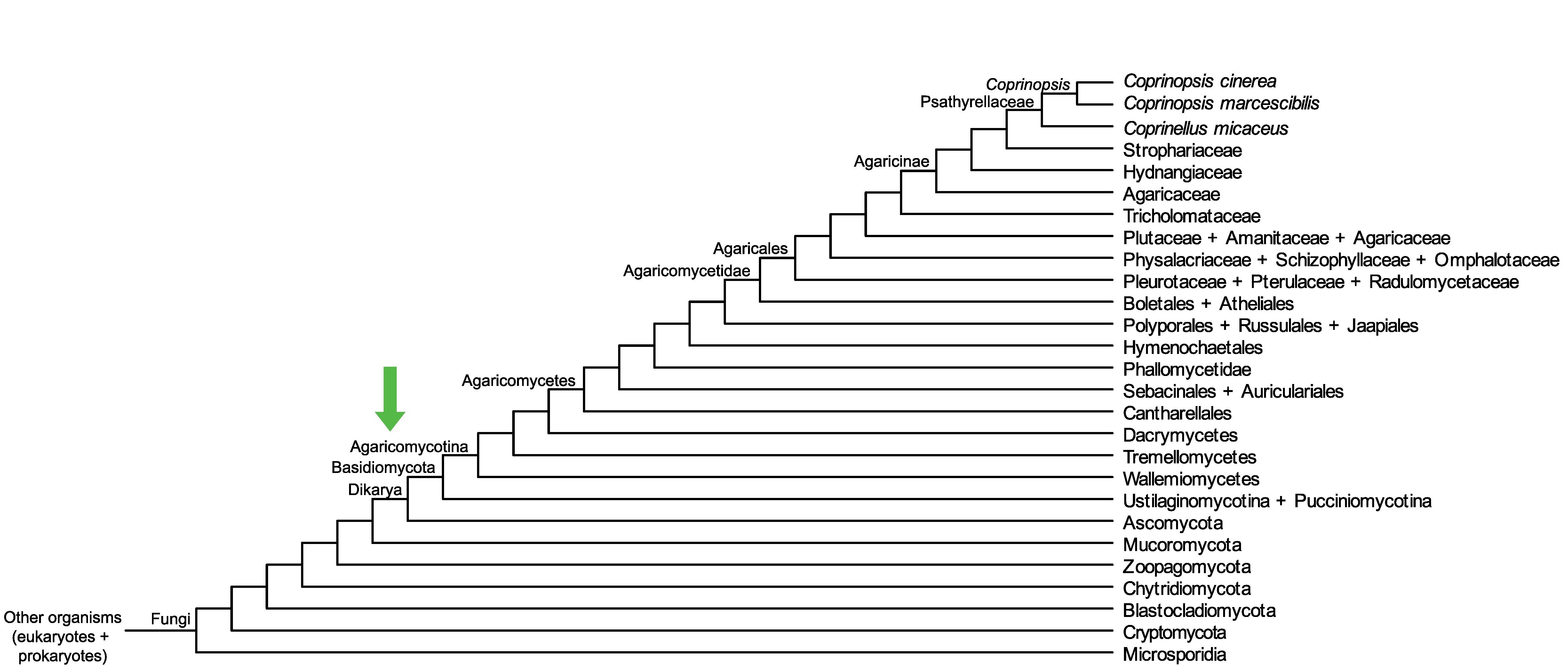
Conservation of CopciAB_361663 across fungi.
Arrow shows the origin of gene family containing CopciAB_361663.
Protein
| Sequence id | CopciAB_361663.T0 |
|---|---|
| Sequence |
>CopciAB_361663.T0 MSQPDVHQLYQRLETIGKGAYGSVHKGKHIPTGNVVALKIINLDTPDDDVADIQREVALLTQLRDAPNITKYFGC YMDGPRVWIVMEYAQGGSVLTLMKASKDGCIEEKYTSVIIREVLVALSYLHKVPVIHRDLKAANILVTATGKVMI CDFGVSALLTTVSSKRTTLTGTPYWMAPEVIQGVSAYDTKADIWSLGIMIFEMIKGTPPHSNLDKFKVMDLIPRA KPPRLQESEGSKDMRDFMSYCLKESPTERLPAEELLKTKWIKSISKVNVSILKDLILRLQQTGPRASLAGPLDWE LEEEADQNAQQDWEFDTVRFDRRPSLDEEDYNAPSQATPTDSFETVRVPASAKTLPSSLRHLFGDSASTEPEPFT VPPPFTRPLTPPQSTTPVTNEWNTIHHVPSADDFASSTSNTLRRADNPPIAEPGEEPQPIISPDVPSPLEASLAP QPIQIPEPPREELQPAFIQPHLGLPAEDLPSFTLSQQTPGRSTPSRLDPPARSKPATHQSTISLDNTSAPGNGNL GVTYSPSPIVRTRSATTLPAIQPPPVSIIPRAFVQQSSPVKSATFTDDPSARGGPGLKDVLKIPSLTTESHLGMV DLLPPSPSALVTSNRSPGAHGISKPSRSYGGPMDYPSPATSKFAMEQDRSQRSAQNSISVDGLPTLEPLNYAALI SNGDPQNELMQTIEGLASWLKVVDQGFQAMLEHSAFGDILEEGNEQYGSSTNESDGQ |
| Length | 732 |
Coding
| Sequence id | CopciAB_361663.T0 |
|---|---|
| Sequence |
>CopciAB_361663.T0 ATGTCCCAGCCAGATGTCCACCAGCTCTACCAGCGTCTAGAAACAATAGGAAAGGGCGCCTATGGTTCAGTCCAC AAGGGAAAACATATTCCAACCGGAAATGTAGTCGCCCTCAAGATCATCAACCTCGACACACCCGACGACGATGTC GCCGACATTCAGCGCGAGGTCGCCCTCCTCACTCAGCTCCGCGACGCCCCCAACATCACAAAGTACTTTGGCTGC TACATGGACGGTCCCCGTGTCTGGATTGTCATGGAGTATGCTCAGGGAGGCAGCGTCCTGACCCTCATGAAAGCC TCCAAAGACGGCTGCATCGAGGAGAAATACACTTCCGTCATCATAAGAGAGGTCCTCGTCGCCCTGAGCTACCTC CACAAGGTCCCAGTCATCCACAGAGACCTCAAGGCAGCCAATATCCTCGTCACCGCTACCGGCAAGGTCATGATC TGTGACTTTGGGGTCTCTGCGCTCCTCACGACTGTCTCCAGCAAACGGACTACCTTGACTGGCACACCGTATTGG ATGGCGCCTGAGGTTATCCAGGGAGTTTCGGCGTATGATACCAAGGCAGACATCTGGAGCTTGGGTATTATGATT TTTGAAATGATCAAGGGGACTCCTCCGCATTCCAACCTGGACAAGTTCAAGGTCATGGACCTGATCCCACGAGCG AAACCACCTCGGCTGCAGGAGAGTGAGGGGAGTAAGGACATGCGCGATTTCATGTCATATTGCTTAAAAGAGTCG CCAACAGAGCGACTCCCGGCAGAAGAATTGTTGAAGACCAAGTGGATCAAGTCGATTTCCAAAGTGAACGTCTCT ATTCTGAAAGATCTCATTCTTCGATTACAACAAACGGGACCGAGAGCCAGTCTTGCTGGCCCGCTGGATTGGGAA TTGGAAGAAGAGGCCGACCAGAATGCGCAACAAGACTGGGAATTTGACACAGTCAGGTTTGACCGGAGACCTTCG CTGGACGAAGAAGACTATAATGCACCATCTCAAGCAACACCTACAGACAGCTTTGAGACCGTCCGTGTACCCGCA TCCGCCAAAACTCTTCCATCCTCACTGCGACATCTATTTGGAGATTCGGCCAGTACCGAGCCAGAACCATTCACT GTCCCTCCGCCATTTACGCGACCACTTACGCCTCCCCAATCCACGACTCCCGTGACCAATGAATGGAATACAATA CACCACGTGCCATCTGCCGACGACTTTGCATCTTCTACATCCAATACCCTTCGCAGGGCAGATAACCCACCAATA GCCGAACCTGGAGAAGAACCGCAACCAATCATTTCTCCCGATGTACCCTCACCACTGGAGGCCTCTTTGGCCCCC CAACCCATCCAAATTCCGGAGCCACCGCGAGAAGAGTTACAACCTGCATTCATCCAACCACACCTTGGCCTCCCT GCTGAGGACTTACCTTCGTTTACACTTTCGCAGCAGACACCTGGCCGATCAACACCATCCCGGCTAGATCCGCCT GCGCGCTCCAAGCCTGCAACTCACCAAAGCACCATATCTTTGGACAATACATCTGCGCCTGGGAACGGAAACCTG GGTGTTACTTATTCTCCTTCGCCCATAGTGCGGACACGGTCGGCGACCACTTTACCTGCCATCCAACCACCGCCC GTTTCTATCATCCCTCGAGCATTTGTTCAGCAATCGTCTCCCGTGAAATCTGCTACGTTCACCGATGATCCAAGT GCACGAGGCGGCCCGGGTTTGAAGGATGTTTTAAAGATCCCTTCACTTACCACTGAGAGTCATCTTGGGATGGTC GATCTTCTGCCCCCGTCACCCTCTGCGCTGGTTACTTCAAATCGCTCGCCTGGGGCACACGGAATATCGAAACCA TCCCGATCTTATGGTGGACCTATGGACTATCCTTCACCGGCTACAAGCAAATTCGCAATGGAGCAAGATCGTTCA CAACGAAGCGCGCAGAACTCGATATCAGTGGATGGTTTGCCGACTTTGGAACCTCTGAACTATGCGGCGTTGATT TCCAACGGGGACCCTCAAAATGAGCTCATGCAGACCATTGAGGGACTGGCCAGTTGGTTGAAGGTGGTTGATCAA GGGTTCCAAGCGATGTTAGAGCATTCGGCTTTTGGAGATATTCTTGAGGAGGGGAATGAGCAGTACGGGTCGTCA ACGAATGAGAGTGATGGGCAGTAA |
| Length | 2199 |
Transcript
| Sequence id | CopciAB_361663.T0 |
|---|---|
| Sequence |
>CopciAB_361663.T0 GACTTCCATGTCCCAGCCAGATGTCCACCAGCTCTACCAGCGTCTAGAAACAATAGGAAAGGGCGCCTATGGTTC AGTCCACAAGGGAAAACATATTCCAACCGGAAATGTAGTCGCCCTCAAGATCATCAACCTCGACACACCCGACGA CGATGTCGCCGACATTCAGCGCGAGGTCGCCCTCCTCACTCAGCTCCGCGACGCCCCCAACATCACAAAGTACTT TGGCTGCTACATGGACGGTCCCCGTGTCTGGATTGTCATGGAGTATGCTCAGGGAGGCAGCGTCCTGACCCTCAT GAAAGCCTCCAAAGACGGCTGCATCGAGGAGAAATACACTTCCGTCATCATAAGAGAGGTCCTCGTCGCCCTGAG CTACCTCCACAAGGTCCCAGTCATCCACAGAGACCTCAAGGCAGCCAATATCCTCGTCACCGCTACCGGCAAGGT CATGATCTGTGACTTTGGGGTCTCTGCGCTCCTCACGACTGTCTCCAGCAAACGGACTACCTTGACTGGCACACC GTATTGGATGGCGCCTGAGGTTATCCAGGGAGTTTCGGCGTATGATACCAAGGCAGACATCTGGAGCTTGGGTAT TATGATTTTTGAAATGATCAAGGGGACTCCTCCGCATTCCAACCTGGACAAGTTCAAGGTCATGGACCTGATCCC ACGAGCGAAACCACCTCGGCTGCAGGAGAGTGAGGGGAGTAAGGACATGCGCGATTTCATGTCATATTGCTTAAA AGAGTCGCCAACAGAGCGACTCCCGGCAGAAGAATTGTTGAAGACCAAGTGGATCAAGTCGATTTCCAAAGTGAA CGTCTCTATTCTGAAAGATCTCATTCTTCGATTACAACAAACGGGACCGAGAGCCAGTCTTGCTGGCCCGCTGGA TTGGGAATTGGAAGAAGAGGCCGACCAGAATGCGCAACAAGACTGGGAATTTGACACAGTCAGGTTTGACCGGAG ACCTTCGCTGGACGAAGAAGACTATAATGCACCATCTCAAGCAACACCTACAGACAGCTTTGAGACCGTCCGTGT ACCCGCATCCGCCAAAACTCTTCCATCCTCACTGCGACATCTATTTGGAGATTCGGCCAGTACCGAGCCAGAACC ATTCACTGTCCCTCCGCCATTTACGCGACCACTTACGCCTCCCCAATCCACGACTCCCGTGACCAATGAATGGAA TACAATACACCACGTGCCATCTGCCGACGACTTTGCATCTTCTACATCCAATACCCTTCGCAGGGCAGATAACCC ACCAATAGCCGAACCTGGAGAAGAACCGCAACCAATCATTTCTCCCGATGTACCCTCACCACTGGAGGCCTCTTT GGCCCCCCAACCCATCCAAATTCCGGAGCCACCGCGAGAAGAGTTACAACCTGCATTCATCCAACCACACCTTGG CCTCCCTGCTGAGGACTTACCTTCGTTTACACTTTCGCAGCAGACACCTGGCCGATCAACACCATCCCGGCTAGA TCCGCCTGCGCGCTCCAAGCCTGCAACTCACCAAAGCACCATATCTTTGGACAATACATCTGCGCCTGGGAACGG AAACCTGGGTGTTACTTATTCTCCTTCGCCCATAGTGCGGACACGGTCGGCGACCACTTTACCTGCCATCCAACC ACCGCCCGTTTCTATCATCCCTCGAGCATTTGTTCAGCAATCGTCTCCCGTGAAATCTGCTACGTTCACCGATGA TCCAAGTGCACGAGGCGGCCCGGGTTTGAAGGATGTTTTAAAGATCCCTTCACTTACCACTGAGAGTCATCTTGG GATGGTCGATCTTCTGCCCCCGTCACCCTCTGCGCTGGTTACTTCAAATCGCTCGCCTGGGGCACACGGAATATC GAAACCATCCCGATCTTATGGTGGACCTATGGACTATCCTTCACCGGCTACAAGCAAATTCGCAATGGAGCAAGA TCGTTCACAACGAAGCGCGCAGAACTCGATATCAGTGGATGGTTTGCCGACTTTGGAACCTCTGAACTATGCGGC GTTGATTTCCAACGGGGACCCTCAAAATGAGCTCATGCAGACCATTGAGGGACTGGCCAGTTGGTTGAAGGTGGT TGATCAAGGGTTCCAAGCGATGTTAGAGCATTCGGCTTTTGGAGATATTCTTGAGGAGGGGAATGAGCAGTACGG GTCGTCAACGAATGAGAGTGATGGGCAGTAAATATGTGTGCTAGTGATACAATTTGGCTCTTTACTTC |
| Length | 2243 |
Gene
| Sequence id | CopciAB_361663.T0 |
|---|---|
| Sequence |
>CopciAB_361663.T0 GACTTCCATGTCCCAGCCAGATGTCCACCAGCTCTACCAGCGTCTAGAAACAATAGGAAAGGGCGCCTATGGTTC AGTCCACAAGGGAAAACATATTCCAACCGGAAATGTAGTCGCCCTCAAGATCATCAACCTCGACACACCCGACGA CGATGTCGCCGACATTCAGCGCGAGGTCGCCCTCCTCACTCAGCTCCGCGACGCCCCCAACATCACAAAGTACTT TGGCTGCTACATGGACGGTCCCCGTGTCTGGATTGTCATGGAGTATGCTCAGGGAGGCAGCGTCCTGACCCTCAT GAAAGCCTCCAAAGACGGCTGCATCGAGGAGAAATACACTTCCGTCATCATAAGAGAGGTCCTCGTCGCCCTGAG CTACCTCCACAAGGTCCCAGTCATCCACAGAGACCTCAAGGCAGCCAATATCCTCGTCACCGCTACCGGCAAGGT CATGATCTGTGACTTTGGGGTCTCTGCGCTCCTCACGACTGTCTCCAGCAAACGGACTACCTTGACTGGCACACC GTATTGGATGGCGCCTGAGGTTATCCAGGGAGTTTCGGCGTATGATACCAAGGCAGACATCTGGAGCTTGGGTAT TATGATTTTTGAAATGATCAAGGGGACTCCTCCGCATTCCAACCTGGACAAGTTCAAGGTCATGGACCTGATCCC ACGAGCGAAACCACCTCGGCTGCAGGAGAGTGAGGGGAGTAAGGACATGCGCGATTTCATGTCATATTGCTTAAA AGAGTCGCCAACAGAGGTACGCGGAGCGCATTTCCTTCGGGAGGATTGCTAATCGCTCTTCAGCGACTCCCGGCA GAAGAATTGTTGAAGACCAAGTGGATCAAGTCGATTTCCAAAGTGAACGTCTCTATTCTGAAAGATCTCATTCTT CGATTACAACAAACGGGACCGAGAGCCAGTCTTGCTGGCCCGCTGGATTGGGAATTGGAAGAAGAGGCCGACCAG TGCGTACATTATTGTGCTTCGCTTGTATTTCTCTTGACGTCTGTCCAGGAATGCGCAACAAGACTGGGAATTTGA CACAGTCAGGTTTGACCGGAGACCTTCGCTGGACGAAGAAGACTATAATGCACCATCTCAAGCAACACCTACAGA CAGCTTTGAGACCGTCCGTGTACCCGCATCCGCCAAAACTCTTCCATCCTCACTGCGACATCTATTTGGAGATTC GGCCAGTACCGAGCCAGAACCATTCACTGTCCCTCCGCCATTTACGCGACCACTTACGCCTCCCCAATCCACGAC TCCCGTGACCAATGAATGGAATACAATACACCACGTGCCATCTGCCGACGACTTTGCATCTTCTACATCCAATAC CCTTCGCAGGGCAGATAACCCACCAATAGCCGAACCTGGAGAAGAACCGCAACCAATCATTTCTCCCGATGTACC CTCACCACTGGAGGCCTCTTTGGCCCCCCAACCCATCCAAATTCCGGAGCCACCGCGAGAAGAGTTACAACCTGC ATTCATCCAACCACACCTTGGCCTCCCTGCTGAGGACTTACCTTCGTTTACACTTTCGCAGCAGACACCTGGCCG ATCAACACCATCCCGGCTAGATCCGCCTGCGCGCTCCAAGCCTGCAACTCACCAAAGCACCATATCTTTGGACAA TACATCTGCGCCTGGGAACGGAAACCTGGGTGTTACTTATTCTCCTTCGCCCATAGTGCGGACACGGTCGGCGAC CACTTTACCTGCCATCCAACCACCGCCCGTTTCTATCATCCCTCGAGCATTTGTTCAGCAATCGTCTCCCGTGAA ATCTGCTACGTTCACCGATGATCCAAGTGCACGAGGCGGCCCGGGTTTGAAGGATGTTTTAAAGGTGAGTTATTC TCTCTCTTATTATATGCATCGCGGCCTCAAGAGTTTGTCAGATCCCTTCACTTACCACTGAGAGTCATCTTGGGA TGGTCGATCTTCTGCCCCCGTCACCCTCTGCGCTGGTTACTTCAAATCGCTCGCCTGGGGCACACGGAATATCGA AACCATCCCGATCTTATGGTGGACCTATGGACTATCCTTCACCGGCTACAAGCAAATTCGCAATGGAGCAAGATC GTTCACAACGAAGCGCGCAGAACTCGATATCAGTGGATGGTTTGCCGACTTTGGAACCTCTGAACTATGCGGCGT TGATTTCCAACGGGGACCCTCAAAATGAGCTCATGCAGACCATTGAGGGACTGGCCAGTTGGTTGAAGGTGGTTG ATCAAGGGTTCCAAGCGATGTTAGAGCATTCGGCTTTTGGAGATATTCTTGAGGAGGGGAATGAGCAGTACGGGT CGTCAACGAATGAGAGTGATGGGCAGTAAATATGTGTGCTAGTGATACAATTTGGCTCTTTACTTC |
| Length | 2391 |