CopciAB_362355
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_362355 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 362355 |
| Uniprot id | Functional description | protein kinase activity | |
| Location | scaffold_3:766923..768789 | Strand | - |
| Gene length (nt) | 1867 | Transcript length (nt) | 1521 |
| CDS length (nt) | 1482 | Protein length (aa) | 493 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB26366 | 46 | 2.07E-77 | 268 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4612 | 33.6 | 5.535E-68 | 240 |
| Lentinula edodes B17 | Lened_B_1_1_1083 | 33.4 | 2.096E-67 | 238 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1072667 | 42.2 | 2.347E-66 | 235 |
| Agrocybe aegerita | Agrae_CAA7263582 | 37.7 | 3.301E-61 | 220 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1402741 | 44.7 | 3.632E-49 | 184 |
| Pleurotus ostreatus PC9 | PleosPC9_1_61673 | 45.1 | 1.387E-47 | 179 |
| Lentinula edodes NBRC 111202 | Lenedo1_1036096 | 41.1 | 1.584E-44 | 170 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_362355.T0 |
| Description | protein kinase activity |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 226 | 490 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 20 | 0.6072 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K16288 |
EggNOG
| COG category | Description |
|---|---|
| K | protein kinase activity |
| L | protein kinase activity |
| T | protein kinase activity |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
Conservation of CopciAB_362355 across fungi.
Arrow shows the origin of gene family containing CopciAB_362355.
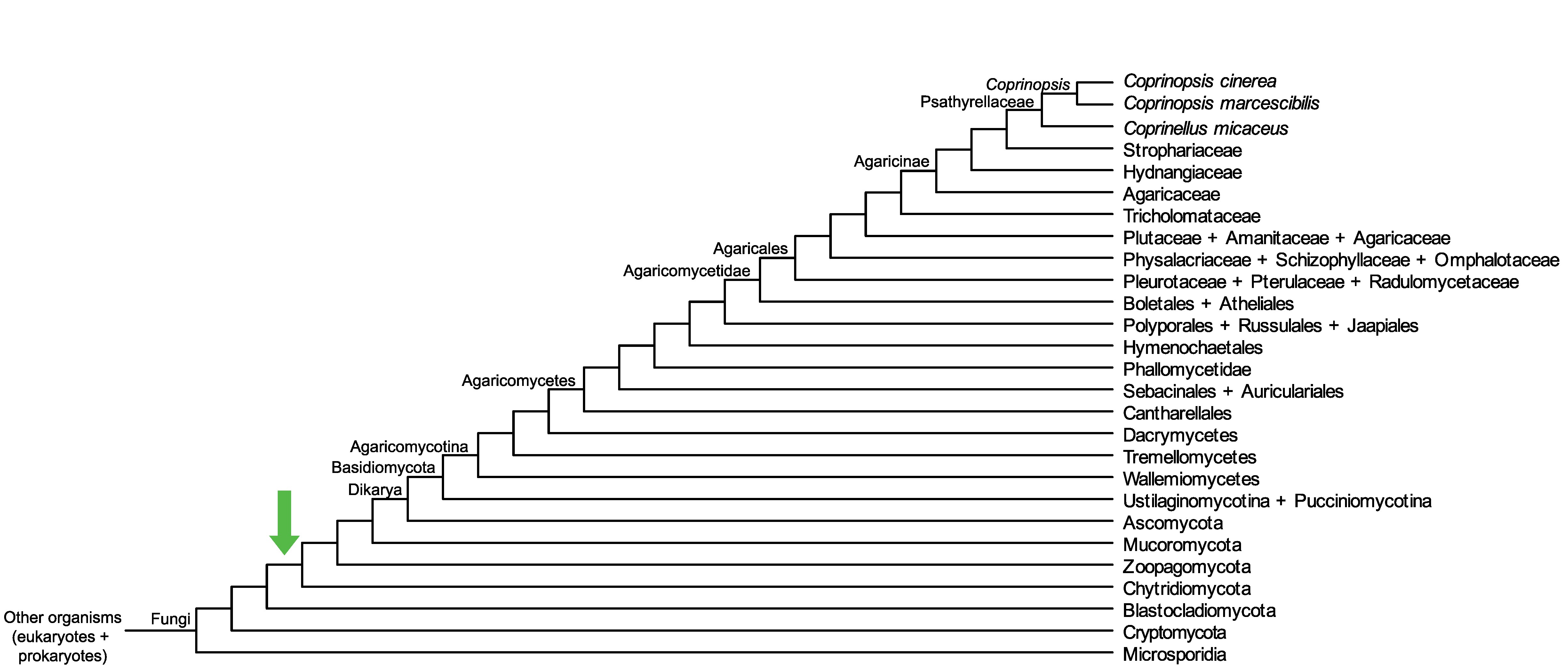
Protein
| Sequence id | CopciAB_362355.T0 |
|---|---|
| Sequence |
>CopciAB_362355.T0 MISTMAAYILAAIASSLGFSASPPAAPLRSSPTLEANQNAERTSAQSKQTAHDSHPAGLSCDSPEVQGDAVDDWD SEDSEPPEDFSSEEEGHGHGEVQMQMEESWSRLGVVPTRRDTGATIRPSRFSPSSPSDSQSESILGHAYSSSSCS GVASRHCYPLRRGSTIDLVTEALDLSAEIEICGGSVAHGGYSDVYRAKWTTTTGSVTGNGNWIVNGDGVSAGAGK GEEVVTRDVAVKTLRAFSQTSNVDLNRARKRLNREVHVLKRLNHPNIAEFYGIAFHKDGRPSVVMKWYRYGTAPE YLAKNAGVDRLGVVRDVAAGLEYLHTLQPPVVHGDLKGSNTLVDESGRVVLSDFGLSKVMEQVLGPTGFTTSTCA GGTVRWLAPELLFDPRMEVDPYDDEDIEAVETPTMASDIWSLGCVAYELATGKYPYADERYDYNVMQAIMNKNRT PADVKEEWTGESRRVYRMLSRCWVREPSNRGNIREIVNLLRDH |
| Length | 493 |
Coding
| Sequence id | CopciAB_362355.T0 |
|---|---|
| Sequence |
>CopciAB_362355.T0 ATGATTTCGACGATGGCGGCGTACATCCTCGCTGCTATAGCCTCCTCTTTGGGGTTCTCTGCATCTCCTCCGGCT GCTCCTCTGCGATCGTCGCCGACCCTCGAGGCCAACCAGAACGCAGAACGTACTAGTGCGCAGTCCAAGCAGACT GCTCACGATTCCCATCCTGCCGGCTTGTCGTGCGATTCCCCTGAGGTCCAGGGTGACGCCGTGGACGATTGGGAT TCAGAGGATTCCGAGCCGCCTGAGGACTTTAGCAGCGAGGAAGAGGGTCACGGTCACGGAGAGGTGCAGATGCAG ATGGAGGAGTCTTGGAGTCGTCTGGGCGTTGTGCCGACGAGGAGAGATACGGGAGCTACCATCCGACCGTCTAGG TTTTCACCCAGTTCACCTTCCGACTCTCAGTCTGAGTCTATCCTTGGCCACGCTTATAGTTCTAGTAGTTGTTCT GGTGTGGCATCGAGACATTGTTATCCTCTTCGACGCGGTTCGACGATCGATTTGGTCACGGAGGCGCTCGACTTG TCGGCAGAGATTGAGATTTGTGGGGGGTCGGTTGCGCATGGTGGGTATTCGGATGTGTATAGAGCCAAGTGGACG ACGACGACGGGGAGTGTGACTGGGAATGGGAATTGGATTGTGAATGGGGACGGGGTTAGTGCTGGTGCTGGTAAA GGAGAGGAGGTTGTGACGCGTGATGTAGCTGTCAAGACTCTCCGGGCGTTCTCGCAGACGTCAAATGTAGACCTC AATCGAGCTCGAAAGCGTCTAAACAGAGAAGTCCACGTCCTGAAACGCCTGAACCACCCCAACATTGCCGAGTTC TACGGAATAGCGTTTCATAAGGACGGGAGACCTTCGGTGGTGATGAAGTGGTATCGGTATGGGACTGCGCCGGAA TATTTGGCGAAGAATGCGGGTGTGGATCGGTTGGGGGTCGTGAGGGATGTAGCAGCTGGATTGGAGTACCTGCAT ACGCTACAGCCGCCTGTCGTGCACGGGGATTTGAAAGGGTCCAATACCCTCGTCGACGAGAGCGGGCGCGTGGTG CTATCAGATTTCGGACTCTCCAAAGTCATGGAACAGGTCCTCGGGCCAACAGGGTTCACGACTTCGACGTGCGCA GGAGGGACAGTCCGGTGGCTGGCCCCCGAGCTACTGTTCGATCCTAGGATGGAGGTGGACCCGTATGACGATGAA GACATTGAAGCCGTCGAGACGCCTACTATGGCATCGGATATTTGGTCTCTTGGATGTGTTGCGTACGAGTTGGCC ACAGGAAAGTACCCGTATGCGGACGAGAGGTACGACTACAACGTGATGCAGGCGATTATGAACAAGAACCGGACC CCAGCTGACGTGAAGGAAGAATGGACGGGAGAGTCTAGGAGGGTCTATAGGATGCTAAGCAGGTGTTGGGTGCGG GAGCCAAGTAATCGAGGTAATATCAGGGAGATTGTTAATTTATTACGAGATCATTAG |
| Length | 1482 |
Transcript
| Sequence id | CopciAB_362355.T0 |
|---|---|
| Sequence |
>CopciAB_362355.T0 GGATGATTTCGACGATGGCGGCGTACATCCTCGCTGCTATAGCCTCCTCTTTGGGGTTCTCTGCATCTCCTCCGG CTGCTCCTCTGCGATCGTCGCCGACCCTCGAGGCCAACCAGAACGCAGAACGTACTAGTGCGCAGTCCAAGCAGA CTGCTCACGATTCCCATCCTGCCGGCTTGTCGTGCGATTCCCCTGAGGTCCAGGGTGACGCCGTGGACGATTGGG ATTCAGAGGATTCCGAGCCGCCTGAGGACTTTAGCAGCGAGGAAGAGGGTCACGGTCACGGAGAGGTGCAGATGC AGATGGAGGAGTCTTGGAGTCGTCTGGGCGTTGTGCCGACGAGGAGAGATACGGGAGCTACCATCCGACCGTCTA GGTTTTCACCCAGTTCACCTTCCGACTCTCAGTCTGAGTCTATCCTTGGCCACGCTTATAGTTCTAGTAGTTGTT CTGGTGTGGCATCGAGACATTGTTATCCTCTTCGACGCGGTTCGACGATCGATTTGGTCACGGAGGCGCTCGACT TGTCGGCAGAGATTGAGATTTGTGGGGGGTCGGTTGCGCATGGTGGGTATTCGGATGTGTATAGAGCCAAGTGGA CGACGACGACGGGGAGTGTGACTGGGAATGGGAATTGGATTGTGAATGGGGACGGGGTTAGTGCTGGTGCTGGTA AAGGAGAGGAGGTTGTGACGCGTGATGTAGCTGTCAAGACTCTCCGGGCGTTCTCGCAGACGTCAAATGTAGACC TCAATCGAGCTCGAAAGCGTCTAAACAGAGAAGTCCACGTCCTGAAACGCCTGAACCACCCCAACATTGCCGAGT TCTACGGAATAGCGTTTCATAAGGACGGGAGACCTTCGGTGGTGATGAAGTGGTATCGGTATGGGACTGCGCCGG AATATTTGGCGAAGAATGCGGGTGTGGATCGGTTGGGGGTCGTGAGGGATGTAGCAGCTGGATTGGAGTACCTGC ATACGCTACAGCCGCCTGTCGTGCACGGGGATTTGAAAGGGTCCAATACCCTCGTCGACGAGAGCGGGCGCGTGG TGCTATCAGATTTCGGACTCTCCAAAGTCATGGAACAGGTCCTCGGGCCAACAGGGTTCACGACTTCGACGTGCG CAGGAGGGACAGTCCGGTGGCTGGCCCCCGAGCTACTGTTCGATCCTAGGATGGAGGTGGACCCGTATGACGATG AAGACATTGAAGCCGTCGAGACGCCTACTATGGCATCGGATATTTGGTCTCTTGGATGTGTTGCGTACGAGTTGG CCACAGGAAAGTACCCGTATGCGGACGAGAGGTACGACTACAACGTGATGCAGGCGATTATGAACAAGAACCGGA CCCCAGCTGACGTGAAGGAAGAATGGACGGGAGAGTCTAGGAGGGTCTATAGGATGCTAAGCAGGTGTTGGGTGC GGGAGCCAAGTAATCGAGGTAATATCAGGGAGATTGTTAATTTATTACGAGATCATTAGTTATCGCGTCAAATAA AAGTCGCGTGGGTTTTGATGA |
| Length | 1521 |
Gene
| Sequence id | CopciAB_362355.T0 |
|---|---|
| Sequence |
>CopciAB_362355.T0 GGATGATTTCGACGATGGCGGCGTACATCCTCGCTGCTATAGCCTCCTCTTTGGGGTTCTCTGCATCTCCTCCGG CTGCTCCTCTGCGATCGTCGCCGACCCTCGAGGCCAACCAGAACGCAGAACGTACTAGTGCGCAGTCCAAGCAGA CTGCTCACGATTCCCATCCTGCCGGCTTGTCGTGCGATTCCCCTGAGGTCCAGGGTGACGCCGTGGACGATTGGG ATTCAGAGGATTCCGAGCCGCCTGAGGACTTTAGCAGCGAGGAAGAGGGTCACGGTCACGGAGAGGTGCAGATGC AGATGGAGGAGTCTTGGAGTCGTCTGGGCGTTGTGCCGACGAGGAGAGATACGGGAGCTACCATCCGACCGTCTA GGTTTTCACCCAGTTCACCTTCCGACTCTCAGTCTGAGTCTATCCTTGGCCACGCTTATAGTTCTAGTAGTTGTT CTGGTGTGGCATCGAGACATTGTTATCCTCTTCGACGTACGTCCGAATTCCCACTGTTCCGTCCGGCGAACGGCT AACTGGATATTGGTGAGATAGGCGGTTCGACGATCGATTTGGTCACGGAGGCGCTCGACTTGTCGGCAGAGATTG AGATTTGTGGGGGGTCGGTTGCGCATGGTGGGTATTCGGATGTGTATAGAGCCAAGTGGACGACGACGACGGGGA GTGTGACTGGGAATGGGAATTGGATTGTGAATGGGGACGGGGTTAGTGCTGGTGCTGGTAAAGGAGAGGAGGTTG TGACGGTGAATACATAGAATCGTAGCTGCGAGTGGCCATCGGGATGTGCGCTGACTTTGGTGTAGCGTGATGTAG CTGTCAAGACTCTCCGGGCGTTCTCGCAGACGTCAAATGTAGACCTCAATCGAGCTCGAAAGGTACTCCCATTCC TTCCCTTCTCGCTCAACCGCTAACCCACTGATCGTCTAACCCCAACTTTGTAGCGTCTAAACAGAGAAGTCCACG TCCTGAAACGCCTGAACCACCCCAACATTGCCGAGTTCTACGGAATAGCGTTTCATAAGGACGGGAGACCTTCGG TGGTGATGAAGTGGTATCGGTATGGGACTGCGCCGGAATATTTGGCGAAGAATGCGGGTGTGGATCGGTTGGGGG TCGTAAGTCGCCTTTACTAGGTTTTAACTAGCCAGAACTGACAGGGGACGTACAGGTGAGGGATGTAGCAGCTGG ATTGGAGTACCTGCATACGCTACAGCCGCCTGTCGTGCACGGGGATTTGAAAGGGGTACGTATCCGAGTCGTTAA CGGTTACCACCATGCAAGGTGACACTGACGTCTGCAGTCCAATACCCTCGTCGACGAGAGCGGGCGCGTGGTGCT ATCAGATTTCGGACTCTCCAAAGTCATGGAACAGGTCCTCGGGCCAACAGGGTTCACGACTTCGACGTGCGCAGG AGGGACAGTCCGGTGGCTGGCCCCCGAGCTACTGTTCGATCCTAGGATGGAGGTGGACCCGTATGACGATGAAGA CATTGAAGCCGTCGAGACGCCTACTATGGCATCGGATATTTGGTCTCTTGGATGTGTTGCGTACGAGGTGAGGGC TAGTCGTGCCTCGAGGCGAGAATAGGGCTGACAGGATACCAGTTGGCCACAGGAAAGTACCCGTATGCGGACGAG AGGTACGACTACAACGTGATGCAGGCGATTATGAACAAGAACCGGACCCCAGCTGACGTGAAGGAAGAATGGACG GGAGAGTCTAGGAGGGTCTATAGGATGCTAAGCAGGTGTTGGGTGCGGGAGCCAAGTAATCGAGGTAATATCAGG GAGATTGTTAATTTATTACGAGATCATTAGTTATCGCGTCAAATAAAAGTCGCGTGGGTTTTGATGA |
| Length | 1867 |