CopciAB_362996
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_362996 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 362996 |
| Uniprot id | Functional description | Aryl-alcohol oxidase | |
| Location | scaffold_10:634969..637560 | Strand | - |
| Gene length (nt) | 2592 | Transcript length (nt) | 2022 |
| CDS length (nt) | 1809 | Protein length (aa) | 602 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7270742 | 62.1 | 2.533E-236 | 735 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1079425 | 53.4 | 1.988E-208 | 654 |
| Pleurotus ostreatus PC9 | PleosPC9_1_88908 | 53.4 | 2.113E-207 | 651 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB27672 | 53.1 | 9.106E-203 | 638 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1466896 | 51 | 9.958E-200 | 629 |
| Flammulina velutipes | Flave_chr06AA00372 | 50.8 | 7.862E-192 | 606 |
| Grifola frondosa | Grifr_OBZ73035 | 47.6 | 8.301E-171 | 545 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_130042 | 44.8 | 3.123E-163 | 523 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_362996.T0 |
| Description | Aryl-alcohol oxidase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 37 | 350 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 457 | 594 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 22 | 0.9709 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| K00108 |
EggNOG
| COG category | Description |
|---|---|
| E | Aryl-alcohol oxidase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
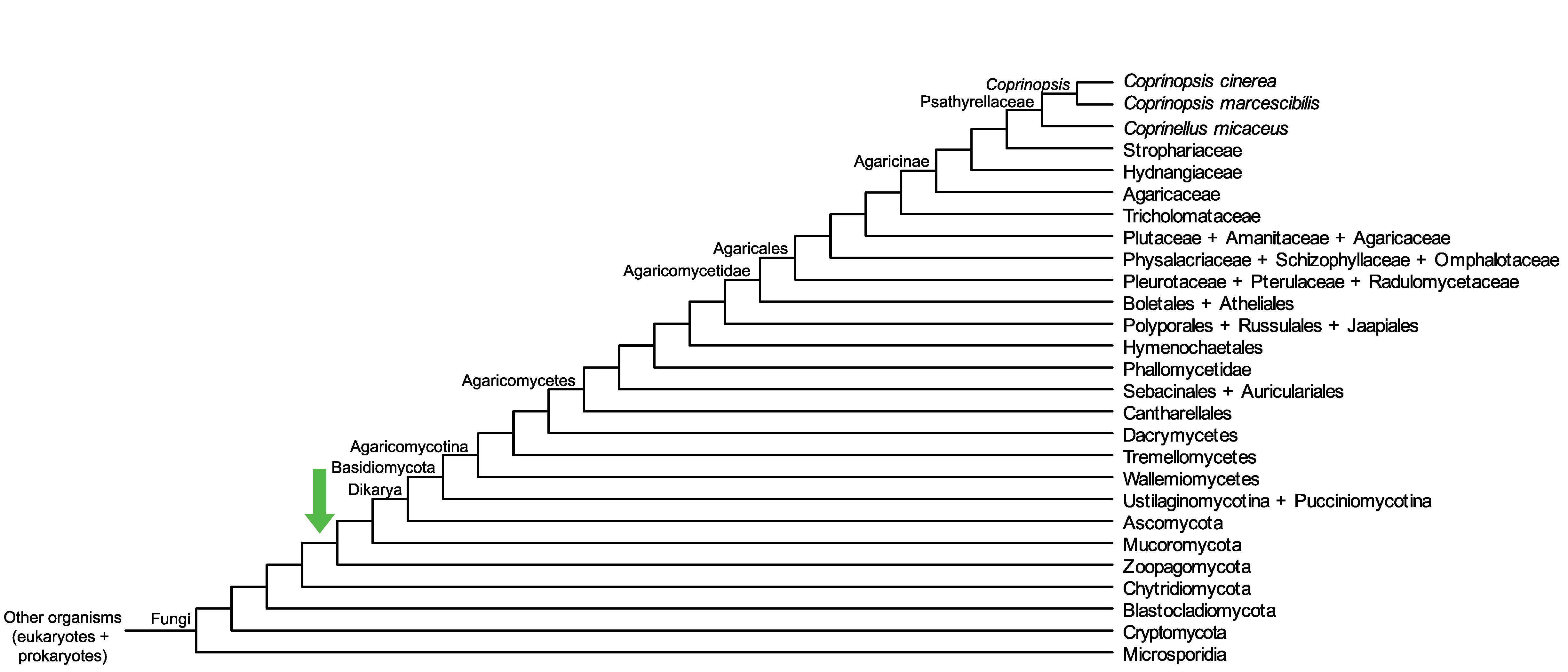
Conservation of CopciAB_362996 across fungi.
Arrow shows the origin of gene family containing CopciAB_362996.
Protein
| Sequence id | CopciAB_362996.T0 |
|---|---|
| Sequence |
>CopciAB_362996.T0 MPTSAGLRLVLSSLLLLPSAFGALLTDPSQLRGDRTYDYVIVGAGNAGNVIAERISAGPRPKSVLVLEAGVSDEG VLAAQVPFLGPTLTPGTPFDWNYTVAPQEGLDGRTFPFPRGKMLGGCSSVNYMVHHFGSSEDYNKLARDSGDNGW SWSSIKKYIFKHEKIVPPADNSDTDGKFLPQFHGTGGTVSVSLPGNSQSIDAKVIATTDELPEFPFNPDQGHGNG QVLGMGWTQNSIGEGARSSSSTTYLKEALKRPHVDVLINAHVTKLVTTRKKRGRPVFDKVQFASGPGAPVTTVTA RREIILSAGAFGTPQILLLSGIGPKTDLDVLGIPTVIHNPSVGQNLSDHVLLPNIFNVRGQDTLDQIIRGDPNVV PGVLDQWTTSRSGPLANGVTNNLGFFRLPANSSIFNSVSDPATGPTASHWEMIVINFYLNPFGPPIPPAGTFMTL ISALISPTSRGFVRLASADPFTAPIIDPKFLTTQFDIFALREAVRATKRFVTASVWNDYVISPWGGLAQTSDEGI DAYVRQQSTTVYHPVGTAAISPKGANYGVVDPDLKLKGAEGVRIADASVWPFLPNAHTQGPVYLLAERAADLILG RA |
| Length | 602 |
Coding
| Sequence id | CopciAB_362996.T0 |
|---|---|
| Sequence |
>CopciAB_362996.T0 ATGCCGACCTCTGCGGGTCTTCGACTCGTTCTCTCGTCGTTGTTGCTGCTTCCTTCTGCGTTCGGCGCTCTCCTC ACCGACCCTTCCCAGTTACGAGGGGATCGTACGTATGACTATGTGATCGTTGGTGCTGGAAACGCAGGCAATGTC ATCGCGGAGAGGATTTCAGCTGGCCCCCGCCCCAAATCCGTGCTCGTTCTCGAAGCTGGCGTCAGCGATGAAGGC GTGCTGGCTGCTCAAGTGCCATTCCTAGGCCCCACTTTGACTCCAGGTACACCCTTCGACTGGAATTACACTGTC GCACCGCAAGAAGGTCTGGATGGGCGTACGTTCCCGTTCCCGCGTGGAAAGATGTTGGGCGGTTGTTCTTCCGTC AATTATATGGTACACCACTTTGGGTCCTCGGAAGACTATAATAAGCTCGCTAGAGACTCCGGTGATAATGGCTGG AGCTGGTCCTCAATTAAAAAGTATATCTTTAAGCACGAGAAGATCGTCCCCCCCGCCGACAACTCAGACACGGAT GGCAAGTTCCTGCCTCAGTTCCATGGGACTGGAGGAACCGTCTCTGTCAGTCTTCCAGGCAACTCGCAGTCGATT GATGCCAAAGTCATCGCGACCACCGATGAACTGCCCGAGTTCCCTTTCAACCCCGATCAAGGACATGGGAATGGT CAGGTCTTGGGAATGGGATGGACACAGAACTCAATTGGCGAGGGAGCTCGGAGCAGTTCCTCGACTACCTACTTG AAGGAAGCACTCAAACGCCCCCATGTCGACGTCTTGATTAACGCGCATGTGACAAAGTTGGTTACTACCCGGAAG AAGAGGGGACGACCAGTGTTTGACAAGGTTCAGTTTGCGAGTGGACCTGGGGCCCCTGTCACGACCGTCACTGCT CGGCGGGAGATCATCCTTTCTGCGGGAGCCTTTGGTACTCCCCAGATCCTGCTATTGTCCGGTATAGGACCGAAG ACAGATCTCGACGTGCTCGGGATCCCAACTGTCATTCACAATCCTTCAGTTGGACAGAACTTGTCTGACCACGTC CTCCTTCCCAACATCTTCAATGTCAGGGGACAAGATACGCTTGACCAGATTATTCGTGGAGACCCGAATGTTGTT CCTGGTGTGCTAGACCAGTGGACTACGTCGAGGAGCGGACCCCTTGCCAACGGTGTTACAAATAACCTTGGGTTC TTCAGGTTGCCAGCGAACTCGAGTATCTTTAATAGCGTGAGCGATCCGGCGACTGGACCTACCGCCTCGCATTGG GAGATGATCGTCATTAACTTCTATCTGAACCCCTTCGGACCTCCCATCCCTCCCGCTGGCACGTTCATGACACTC ATCTCGGCGCTCATCTCTCCCACTTCTCGCGGTTTCGTCCGACTGGCTTCTGCAGACCCCTTCACGGCGCCCATC ATCGACCCCAAGTTCCTCACGACCCAGTTCGATATCTTCGCTTTACGTGAAGCGGTGCGAGCGACCAAACGGTTC GTTACTGCGAGTGTGTGGAATGATTATGTGATTAGCCCGTGGGGTGGGTTGGCGCAGACGAGCGACGAGGGGATC GATGCGTATGTGAGGCAGCAGTCGACGACAGTTTATCATCCTGTTGGTACTGCGGCTATTTCGCCTAAGGGAGCA AACTATGGAGTTGTTGATCCTGATTTGAAGTTGAAGGGAGCGGAGGGTGTTAGGATTGCGGATGCTTCGGTTTGG CCTTTCCTGCCGAATGCGCATACGCAGGGACCGGTGTATCTTCTTGCGGAGAGGGCTGCTGATTTGATTCTTGGG AGAGCTTGA |
| Length | 1809 |
Transcript
| Sequence id | CopciAB_362996.T0 |
|---|---|
| Sequence |
>CopciAB_362996.T0 ATCCCCGCTCCTCTCTCCTCCCTGTCCTCACATTCGATCTGCCGTTCGTTCCATTATGCCGACCTCTGCGGGTCT TCGACTCGTTCTCTCGTCGTTGTTGCTGCTTCCTTCTGCGTTCGGCGCTCTCCTCACCGACCCTTCCCAGTTACG AGGGGATCGTACGTATGACTATGTGATCGTTGGTGCTGGAAACGCAGGCAATGTCATCGCGGAGAGGATTTCAGC TGGCCCCCGCCCCAAATCCGTGCTCGTTCTCGAAGCTGGCGTCAGCGATGAAGGCGTGCTGGCTGCTCAAGTGCC ATTCCTAGGCCCCACTTTGACTCCAGGTACACCCTTCGACTGGAATTACACTGTCGCACCGCAAGAAGGTCTGGA TGGGCGTACGTTCCCGTTCCCGCGTGGAAAGATGTTGGGCGGTTGTTCTTCCGTCAATTATATGGTACACCACTT TGGGTCCTCGGAAGACTATAATAAGCTCGCTAGAGACTCCGGTGATAATGGCTGGAGCTGGTCCTCAATTAAAAA GTATATCTTTAAGCACGAGAAGATCGTCCCCCCCGCCGACAACTCAGACACGGATGGCAAGTTCCTGCCTCAGTT CCATGGGACTGGAGGAACCGTCTCTGTCAGTCTTCCAGGCAACTCGCAGTCGATTGATGCCAAAGTCATCGCGAC CACCGATGAACTGCCCGAGTTCCCTTTCAACCCCGATCAAGGACATGGGAATGGTCAGGTCTTGGGAATGGGATG GACACAGAACTCAATTGGCGAGGGAGCTCGGAGCAGTTCCTCGACTACCTACTTGAAGGAAGCACTCAAACGCCC CCATGTCGACGTCTTGATTAACGCGCATGTGACAAAGTTGGTTACTACCCGGAAGAAGAGGGGACGACCAGTGTT TGACAAGGTTCAGTTTGCGAGTGGACCTGGGGCCCCTGTCACGACCGTCACTGCTCGGCGGGAGATCATCCTTTC TGCGGGAGCCTTTGGTACTCCCCAGATCCTGCTATTGTCCGGTATAGGACCGAAGACAGATCTCGACGTGCTCGG GATCCCAACTGTCATTCACAATCCTTCAGTTGGACAGAACTTGTCTGACCACGTCCTCCTTCCCAACATCTTCAA TGTCAGGGGACAAGATACGCTTGACCAGATTATTCGTGGAGACCCGAATGTTGTTCCTGGTGTGCTAGACCAGTG GACTACGTCGAGGAGCGGACCCCTTGCCAACGGTGTTACAAATAACCTTGGGTTCTTCAGGTTGCCAGCGAACTC GAGTATCTTTAATAGCGTGAGCGATCCGGCGACTGGACCTACCGCCTCGCATTGGGAGATGATCGTCATTAACTT CTATCTGAACCCCTTCGGACCTCCCATCCCTCCCGCTGGCACGTTCATGACACTCATCTCGGCGCTCATCTCTCC CACTTCTCGCGGTTTCGTCCGACTGGCTTCTGCAGACCCCTTCACGGCGCCCATCATCGACCCCAAGTTCCTCAC GACCCAGTTCGATATCTTCGCTTTACGTGAAGCGGTGCGAGCGACCAAACGGTTCGTTACTGCGAGTGTGTGGAA TGATTATGTGATTAGCCCGTGGGGTGGGTTGGCGCAGACGAGCGACGAGGGGATCGATGCGTATGTGAGGCAGCA GTCGACGACAGTTTATCATCCTGTTGGTACTGCGGCTATTTCGCCTAAGGGAGCAAACTATGGAGTTGTTGATCC TGATTTGAAGTTGAAGGGAGCGGAGGGTGTTAGGATTGCGGATGCTTCGGTTTGGCCTTTCCTGCCGAATGCGCA TACGCAGGGACCGGTGTATCTTCTTGCGGAGAGGGCTGCTGATTTGATTCTTGGGAGAGCTTGATTTGGATGCTT TGGATACCCCCAAGTTGTCGTCTTTGAAAGACGTCGTTCGCTCGTTGACTTCTGTCTGGAATTCTAATGGACAGC ACACCACTAATACCTTTCCTGTATAAGATTGTACAACACTTGCTATTGAAATAATCATTCGTCTCTTCAATG |
| Length | 2022 |
Gene
| Sequence id | CopciAB_362996.T0 |
|---|---|
| Sequence |
>CopciAB_362996.T0 ATCCCCGCTCCTCTCTCCTCCCTGTCCTCACATTCGATCTGCCGTTCGTTCCATTATGCCGACCTCTGCGGGTCT TCGACTCGTTCTCTCGTCGTTGTTGCTGCTTCCTTCTGCGTTCGGCGCTCTCCTCACCGACCCTTCCCAGTTACG AGGGGATCGTACGTATGACTATGTGATCGTTGGTGGTGAGTAACTTCATTTTTCCTGTTCTGTAAGACTTCAGCT TGACATTCTTCGTCGCTCATTTATACGTCCTTCGTAGCTGGAAACGCAGGCAATGTCATCGCGGAGAGGATTTCA GCTGGCCCCCGCCCCAAATCCGTGCTCGTTCTCGAAGCTGGCGTCAGGTAAGTCCCGATTTGGCCGTATGTCATC CTCCACAGTGAGTTAATAATAACTATCTGATTAGCGATGAAGGCGTGCTGGCTGCTCAAGTGCCATTCCTAGGCC CCACTTTGACTCCAGGCACGTTCAGGCGTAACTTACTTGGAGGGGCTGTCGTAACTTGCTGACTCTCTTTACCTT ATGGCACTCTAACAGGTACACCCTTCGACTGGAATTACACTGTCGCACCGCAAGAAGGTCTGGATGGGCGTACGT TCCCGTTCCCGCGTGGAAAGATGTTGGGCGGTTGTTCTTCCGTCAGTGAGTTGGGCTTCTTGTTGTGATTGTAAT CTCGATTCTGATTGTGCTGAATGGATGCCTTGTAGATTATATGGTACACCACTTTGGGTCCTCGGAAGACTATAA TAAGCTCGCTAGAGACTCCGGTGATAATGGCTGGAGCTGGTCCTCAATTAAAAAGTATATCTTTAAGGTATGGTG TTTCTCCTGCCCGTTAAGCTTGCAATGCATCTGAAACCCGAAATCTGATCACTTGCTTTCCTCTTTGCAGCACGA GAAGATCGTCCCCCCCGCCGACAACTCAGACACGGATGGCAAGTTCCTGCCTCAGTTCCATGGGACTGGAGGAAC CGTCTCTGTCAGTCTTCCAGGCAACTCGCAGTCGATTGATGCCAAAGTCATCGCGACCACCGATGAACTGCCCGA GTTCCCTTTCAACCCCGATCAAGGACATGGGAATGGTCAGGTCTTGGGAATGGGATGGACACAGAACTCAATTGG CGAGGGAGCTCGGAGCAGTTCCTCGACTACCTACTTGAAGGAAGCACTCAAACGCCCCCATGTCGACGTCTTGAT TAACGCGCATGTGACAAAGTTGGTTACTACCCGGAAGAAGAGGGGACGACCAGTGTTTGACAAGGTTCAGTTTGC GAGTGGACCTGGGGGTGAGTGTTCCCATCAATGGTGTGCAGCAGCAACATTGTCTGATTCCTATTTGATAGCCCC TGTCACGACCGTCACTGCTCGGCGGGAGATCATCCTTTCTGCGGGAGCCTTTGGTACTCCCCAGATCCTGCTATT GTCCGGTATAGGACCGAAGACAGATCTCGACGTGCTCGGGATCCCAACTGTCATTCACAATCCTTCAGTTGGACA GAACTTGTCTGACCACGTCCTCCTTCCCAACATCTTCAATGTCAGGGGACAAGATACGCTTGACCAGATTATTCG TGGAGACCCGAATGTTGTTCCTGGTGTGCTAGACCAGTGGACTACGTCGAGGAGCGGACCCCTTGCCAACGGTGT TACAAATAACCTTGGGTTCTTCAGGTTGCCAGCGAACTCGAGTATCTTTAATAGCGTGAGCGATCCGGCGACTGG ACCTACCGCCTCGCATTGGGAGATGATCGTCATTGTGAGTATCCCGGGTCTTCGACCTGTTGCCCTTGTGTTGAT GATTTCGTAGAACTTCTATCTGAACCCCTTCGGACCTCCCATCCCTCCCGCTGGCACGTTCATGACACTCATCTC GGCGCTCATCTCTCCCACTTCTCGTACGACATCTTCCCTTCCAACCTACTCCCAATTCTAACTGCTCCCATTTGC AGGCGGTTTCGTCCGACTGGCTTCTGCAGACCCCTTCACGGCGCCCATCATCGACCCCAAGTTCCTCACGACCCA GTTCGATATCTTCGCTTTACGTGAAGCGGTGCGAGCGACCAAACGGTTCGTTACTGCGAGTGTGTGGAATGATTA TGTGATTAGCCCGTGGGGTGGGTTGGCGCAGACGAGCGACGAGGGGATCGATGCGTATGTGAGGCAGCAGTCGAC GACAGTTTATCATCCTGTTGGTACTGCGGCTATTTCGCCTAAGGGAGCAAACTATGGAGTTGTTGATCCTGATTT GAAGTTGAAGGGAGCGGAGGGTGTTAGGATTGCGGATGCTTCGGTTTGGGTGAGTTTGGATTGTGTGTTTTGGTG GACTGTGTGCTGATTTGCTTTGTAGCCTTTCCTGCCGAATGCGCATACGCAGGGACCGGTGTATCTTCTTGCGGA GAGGGCTGCTGATTTGATTCTTGGGAGAGCTTGATTTGGATGCTTTGGATACCCCCAAGTTGTCGTCTTTGAAAG ACGTCGTTCGCTCGTTGACTTCTGTCTGGAATTCTAATGGACAGCACACCACTAATACCTTTCCTGTATAAGATT GTACAACACTTGCTATTGAAATAATCATTCGTCTCTTCAATG |
| Length | 2592 |