CopciAB_366605
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_366605 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 366605 |
| Uniprot id | Functional description | Belongs to the GMC oxidoreductase family | |
| Location | scaffold_3:2406979..2409833 | Strand | - |
| Gene length (nt) | 2855 | Transcript length (nt) | 2031 |
| CDS length (nt) | 1812 | Protein length (aa) | 603 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25919 | 53.2 | 4.531E-212 | 665 |
| Agrocybe aegerita | Agrae_CAA7269592 | 53.2 | 6.595E-206 | 647 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191438 | 52.3 | 3.173E-201 | 633 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114404 | 52.5 | 3.306E-201 | 633 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1067243 | 52.4 | 4.322E-199 | 627 |
| Pleurotus ostreatus PC9 | PleosPC9_1_86458 | 51.6 | 1.193E-195 | 617 |
| Lentinula edodes NBRC 111202 | Lenedo1_506227 | 51.2 | 8.185E-193 | 609 |
| Schizophyllum commune H4-8 | Schco3_2643025 | 49.1 | 3.317E-186 | 590 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_15867 | 48 | 6.945E-185 | 586 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1435398 | 47.1 | 2.919E-176 | 561 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_130292 | 48.5 | 1.404E-174 | 556 |
| Flammulina velutipes | Flave_chr11AA00287 | 48 | 1.526E-173 | 553 |
| Grifola frondosa | Grifr_OBZ66993 | 44.4 | 6.779E-153 | 493 |
| Auricularia subglabra | Aurde3_1_1278824 | 43.6 | 3.052E-150 | 486 |
| Lentinula edodes B17 | Lened_B_1_1_905 | 35.1 | 2.514E-104 | 351 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_366605.T0 |
| Description | Belongs to the GMC oxidoreductase family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 2 | 316 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 427 | 589 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| K17066 |
EggNOG
| COG category | Description |
|---|---|
| E | Belongs to the GMC oxidoreductase family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_3 |
Transcription factor
| Group |
|---|
| No records |
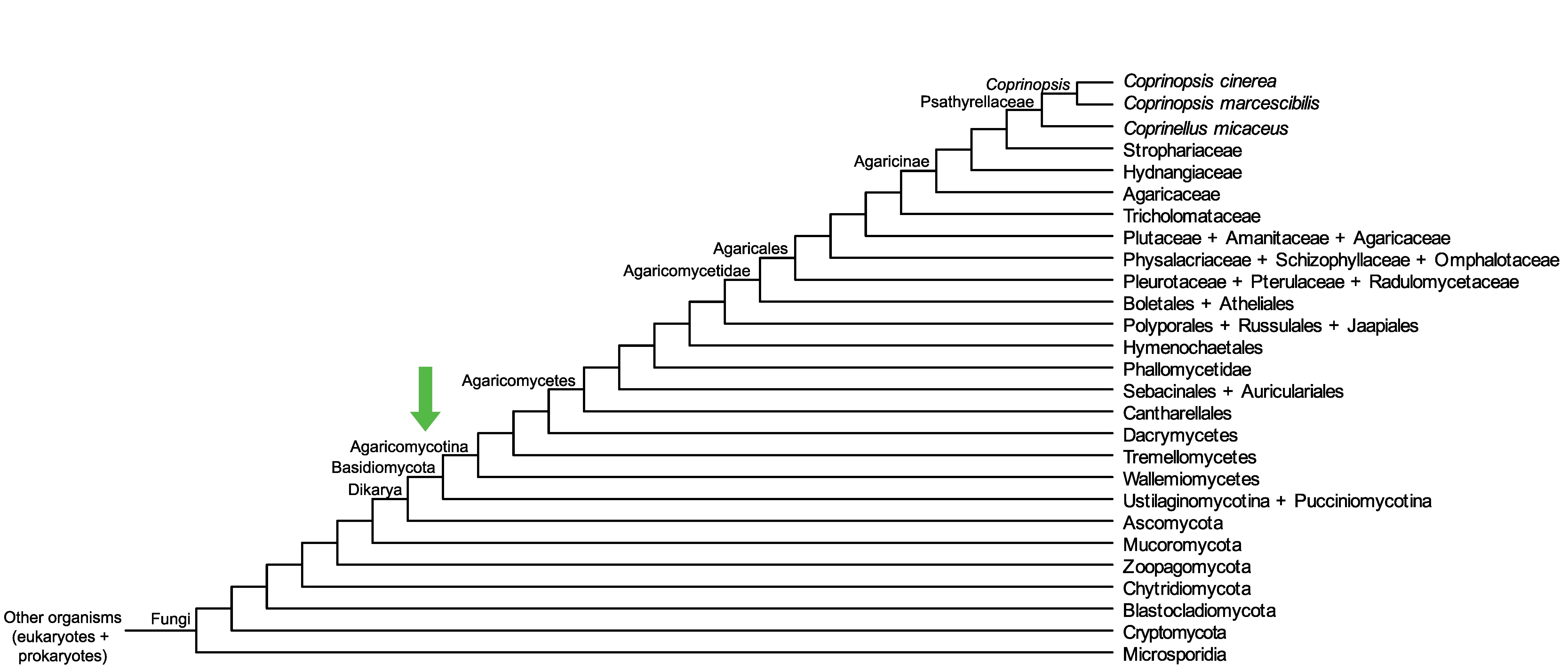
Conservation of CopciAB_366605 across fungi.
Arrow shows the origin of gene family containing CopciAB_366605.
Protein
| Sequence id | CopciAB_366605.T0 |
|---|---|
| Sequence |
>CopciAB_366605.T0 MYDIIVVGGGAAGCVIAGKLSAADPSLKILILEAGPHSHNVPQHVQPARFIRNVQQPQLMDPKSICLHVAQPSEA LLGRKIVMRQGSVLGGGSSVNFMVYVRPSASDYDAWEVQFGNKGWGSKDLIPLLKEVEAYHPFEEGAPVGPVHGD KGPVKISFAKKGNNVGEQFLDVVRQYDKARPIGDWNDFGSGNKYGRWAKYIDPRTGRRSDTAHAFIYNQPPNPNL VVETEKRVVRVLFEGTKAVGVEYVGEEKNEDGSRVVQQAYASKLVVISAGSIASPAILERSGIGGAAVLKANDIA QLVDLPGVGENYMDHSAVFSVYRSAEEADTMDAVYYGGPNELSSYESEWKSSGTGPLSRNGLDAGCKLRPTPDEL KELGPAFQQRWDTFYKDYPDKPVMLGASMAACGSPAGVKEKMWTAAYYSAYPSSTGCVHIAGGRDPFAPLDYHPG YLDKEEDVAMFRWAYKHLREIARRMPFYRGEYEERHPKFPEGSAAACNLSREPVAIDAPKLVYTKEDDEAIDRYH RTTMGPAWHPAGTCAMKPREQGGVLDSRLNVYGTTNLKVADVSVCPENVGANTYSTALAVGLKATVIIAEELGIK LRN |
| Length | 603 |
Coding
| Sequence id | CopciAB_366605.T0 |
|---|---|
| Sequence |
>CopciAB_366605.T0 ATGTACGACATTATCGTCGTTGGCGGTGGTGCCGCTGGATGTGTGATTGCTGGAAAGCTCTCGGCCGCCGACCCA TCGCTGAAGATTCTGATACTCGAAGCCGGACCTCACTCGCACAATGTTCCACAACACGTCCAGCCCGCTCGCTTC ATTCGTAACGTCCAGCAACCCCAGCTCATGGACCCCAAGTCTATCTGCCTTCACGTCGCGCAACCCAGCGAGGCT CTACTGGGCCGCAAAATCGTGATGCGGCAGGGAAGCGTCCTCGGAGGTGGCTCGAGTGTCAATTTCATGGTTTAT GTCCGACCATCAGCTTCGGACTATGACGCATGGGAAGTTCAGTTCGGGAACAAAGGATGGGGCTCGAAAGATCTC ATTCCGCTTCTGAAGGAAGTGGAGGCTTACCATCCTTTCGAGGAGGGTGCTCCGGTTGGGCCAGTCCATGGGGAC AAGGGCCCCGTCAAGATCTCCTTTGCAAAGAAGGGGAATAACGTGGGCGAACAGTTCTTGGACGTCGTCAGGCAG TACGACAAGGCACGTCCGATTGGCGACTGGAACGACTTTGGCTCTGGTAACAAGTACGGGAGATGGGCAAAATAC ATTGATCCAAGGACAGGAAGGCGTTCGGATACTGCTCACGCCTTCATCTACAACCAACCGCCCAACCCCAACCTC GTCGTCGAAACAGAGAAACGCGTCGTACGAGTCTTATTCGAAGGTACCAAAGCCGTAGGCGTGGAATACGTTGGG GAAGAGAAGAACGAGGATGGATCACGAGTCGTCCAACAAGCTTACGCTTCCAAGCTCGTCGTCATTTCCGCTGGA TCCATTGCGTCACCAGCTATTCTCGAGCGCTCCGGCATTGGCGGAGCAGCCGTCTTGAAAGCTAATGACATTGCT CAGCTCGTGGACCTCCCTGGAGTTGGCGAGAACTATATGGACCATTCCGCTGTATTCTCCGTTTATCGTTCTGCC GAAGAAGCTGATACCATGGACGCTGTCTATTACGGTGGCCCGAACGAATTGTCTTCCTATGAATCCGAATGGAAA TCGTCAGGCACAGGCCCACTCTCCCGAAATGGCTTGGACGCAGGCTGCAAGTTGCGTCCGACACCAGATGAACTG AAGGAGCTCGGGCCAGCGTTCCAACAAAGATGGGATACCTTCTACAAGGACTACCCCGACAAACCGGTCATGCTT GGTGCGAGTATGGCCGCTTGTGGTTCGCCCGCGGGTGTCAAGGAGAAGATGTGGACAGCTGCGTATTACTCCGCA TACCCATCCTCGACTGGGTGTGTCCATATCGCCGGAGGACGCGACCCATTCGCTCCCTTGGACTACCATCCGGGG TATCTCGACAAAGAGGAAGACGTAGCCATGTTCCGATGGGCGTACAAACACTTGAGGGAGATAGCTCGACGCATG CCGTTCTACCGTGGCGAATACGAGGAGCGTCACCCCAAGTTTCCCGAAGGAAGTGCTGCCGCGTGTAACCTGTCT AGGGAACCCGTCGCAATCGACGCTCCCAAGTTGGTTTACACGAAGGAAGATGATGAGGCGATCGATAGGTACCAT CGGACCACCATGGGACCAGCTTGGCATCCGGCCGGTACTTGTGCGATGAAGCCTCGAGAGCAAGGCGGCGTCCTC GATTCCAGACTCAACGTCTATGGAACCACCAATCTGAAGGTTGCCGATGTCAGTGTCTGTCCCGAGAACGTCGGA GCCAACACGTACAGTACGGCTTTGGCGGTTGGTCTCAAGGCCACTGTGATCATCGCGGAGGAGCTGGGGATCAAG CTGAGGAACTGA |
| Length | 1812 |
Transcript
| Sequence id | CopciAB_366605.T0 |
|---|---|
| Sequence |
>CopciAB_366605.T0 ACCGACAGGAGGTCCACCTTTGCGACCTATCTTTGACCTCTCTTTCACTCACCTCTCTGACTATGTACGACATTA TCGTCGTTGGCGGTGGTGCCGCTGGATGTGTGATTGCTGGAAAGCTCTCGGCCGCCGACCCATCGCTGAAGATTC TGATACTCGAAGCCGGACCTCACTCGCACAATGTTCCACAACACGTCCAGCCCGCTCGCTTCATTCGTAACGTCC AGCAACCCCAGCTCATGGACCCCAAGTCTATCTGCCTTCACGTCGCGCAACCCAGCGAGGCTCTACTGGGCCGCA AAATCGTGATGCGGCAGGGAAGCGTCCTCGGAGGTGGCTCGAGTGTCAATTTCATGGTTTATGTCCGACCATCAG CTTCGGACTATGACGCATGGGAAGTTCAGTTCGGGAACAAAGGATGGGGCTCGAAAGATCTCATTCCGCTTCTGA AGGAAGTGGAGGCTTACCATCCTTTCGAGGAGGGTGCTCCGGTTGGGCCAGTCCATGGGGACAAGGGCCCCGTCA AGATCTCCTTTGCAAAGAAGGGGAATAACGTGGGCGAACAGTTCTTGGACGTCGTCAGGCAGTACGACAAGGCAC GTCCGATTGGCGACTGGAACGACTTTGGCTCTGGTAACAAGTACGGGAGATGGGCAAAATACATTGATCCAAGGA CAGGAAGGCGTTCGGATACTGCTCACGCCTTCATCTACAACCAACCGCCCAACCCCAACCTCGTCGTCGAAACAG AGAAACGCGTCGTACGAGTCTTATTCGAAGGTACCAAAGCCGTAGGCGTGGAATACGTTGGGGAAGAGAAGAACG AGGATGGATCACGAGTCGTCCAACAAGCTTACGCTTCCAAGCTCGTCGTCATTTCCGCTGGATCCATTGCGTCAC CAGCTATTCTCGAGCGCTCCGGCATTGGCGGAGCAGCCGTCTTGAAAGCTAATGACATTGCTCAGCTCGTGGACC TCCCTGGAGTTGGCGAGAACTATATGGACCATTCCGCTGTATTCTCCGTTTATCGTTCTGCCGAAGAAGCTGATA CCATGGACGCTGTCTATTACGGTGGCCCGAACGAATTGTCTTCCTATGAATCCGAATGGAAATCGTCAGGCACAG GCCCACTCTCCCGAAATGGCTTGGACGCAGGCTGCAAGTTGCGTCCGACACCAGATGAACTGAAGGAGCTCGGGC CAGCGTTCCAACAAAGATGGGATACCTTCTACAAGGACTACCCCGACAAACCGGTCATGCTTGGTGCGAGTATGG CCGCTTGTGGTTCGCCCGCGGGTGTCAAGGAGAAGATGTGGACAGCTGCGTATTACTCCGCATACCCATCCTCGA CTGGGTGTGTCCATATCGCCGGAGGACGCGACCCATTCGCTCCCTTGGACTACCATCCGGGGTATCTCGACAAAG AGGAAGACGTAGCCATGTTCCGATGGGCGTACAAACACTTGAGGGAGATAGCTCGACGCATGCCGTTCTACCGTG GCGAATACGAGGAGCGTCACCCCAAGTTTCCCGAAGGAAGTGCTGCCGCGTGTAACCTGTCTAGGGAACCCGTCG CAATCGACGCTCCCAAGTTGGTTTACACGAAGGAAGATGATGAGGCGATCGATAGGTACCATCGGACCACCATGG GACCAGCTTGGCATCCGGCCGGTACTTGTGCGATGAAGCCTCGAGAGCAAGGCGGCGTCCTCGATTCCAGACTCA ACGTCTATGGAACCACCAATCTGAAGGTTGCCGATGTCAGTGTCTGTCCCGAGAACGTCGGAGCCAACACGTACA GTACGGCTTTGGCGGTTGGTCTCAAGGCCACTGTGATCATCGCGGAGGAGCTGGGGATCAAGCTGAGGAACTGAG TGCAGGACATTGGCTGGTAGTCATCCTCTTTTTTTTCAAGTGTGATAAACAATTATGTATACCTCAGCTACTATG TACTGGTACTCGTTGATGTAAATTAGCCTTAGTCTGAGTGAGCGGACTCACCTCGGATATTTAAGCGCCCATGAC TCAACC |
| Length | 2031 |
Gene
| Sequence id | CopciAB_366605.T0 |
|---|---|
| Sequence |
>CopciAB_366605.T0 ACCGACAGGAGGTCCACCTTTGCGACCTATCTTTGACCTCTCTTTCACTCACCTCTCTGACTATGTACGACATTA TCGTCGTTGGCGGTACGTTCCGCCCATTTTCGCCCCAAGTGTGACGCTGACACACCTCACGCTCACCAGGTGGTG CCGCTGGATGTGTGATTGCTGGAAAGCTCTCGGCCGCCGACCCATCGCTGAAGATTCTGGTAGCATCGAACGCAC GCGCTACTTCAGAGTAGCTTACTAACTCAGCGGCTGCACCATAGATACTCGAAGCCGGACCTCACTCGCACAATG TTCCACAACACGTCCAGCCCGCTCGCTTCATTCGTAACGTCCAGCAACCCCAGCTCATGGACCCCAAGTCTATCT GCCTTCACGTCGCGCAACCCAGCGAGGCTCTACTGGGCCGCAAAATCGTGATGCGGCAGGGAAGCGTCCTCGGAG GTGGCTCGAGTGTCAATTGTGAGTCTCCACCCTCCACCAAGGTTTGTTCCGACGCTGATAGCCTTCACAGTCATG GTTTATGTCCGACCATCAGCTTCGGACTATGACGCATGGGAAGTTCAGTTCGGGAACAAAGGATGGGGCTCGAAA GATCTCATTCCGCTTCTGAAGGAAGTGGAGGCTTACCATCCTTTCGAGGAGGGTGCTCCGGTTGGGCCAGTCCAT GGGGACAAGGGCCCCGTCAAGATCTCCTTTGCAAAGAAGGGGAATAACGTGGGCGAACAGTTCTTGGACGTCGTC AGGCAGTACGACAAGGCACGTCCGATTGGCGACTGGAACGACTTTGGCTCTGGTAACAAGTACGGGGTGAGTGCT TGATATCCTTTCGGCCTCGATGCTTTCTCAGCTGCGTTGGTTACGAACAGAGATGGGCAAAGTAAGCATACTGGC TGTATCCTCGGAATTCAAGGTTAACTCGTTGAGTAGATACATTGATCCAAGGACAGGAAGGCGTTCGGATACTGC TCACGCCTTCATCTACAACCAACCGCCCAACCCCAACCTCGTCGTCGAAACAGAGAAACGCGTCGTACGAGTCTT ATTCGAGTATGTTTCCCTACTGGGTTTGCATCCCCTAACACATAATGAACTATTTTCGCCTCGCACTTTAGAGGT ACCAAAGCCGTAGGCGTGGAATACGTTGGGGAAGAGAAGAACGAGGATGGATCACGAGTCGTCCAACAAGCTTAC GCTTCCAAGCTCGTCGTCATTTCCGCTGGATCCATTGCGTCACCAGCTATTCTCGAGCGCTCCGGCATTGGCGGA GCAGCCGTCTTGAAAGCTAATGACATTGCTCAGCTCGTGGACCTCCCTGGAGTTGGCGAGAACTATATGGGTAAC AACGTCGCTTGGAGGATACAACTGTCATAACTGACACGCATTTTCAGACCATTCCGCTGTATTCTCCGTTTATCG TTCTGCCGAAGAAGCTGATACCATGGACGCTGTCTATTACGGTGGCCCGAACGAATTGTCTTGTGAGTGAACTGT CCATTAACCTCAGGATAACCGGGTTCATCTTTCGTCTTGCCAGCCTATGAATCCGAATGGAAATCGTCAGGCACA GGCCCACTCTCCCGAAAGTAAGGCCCACTTTATCCTCTCTTCACGTATCTGCTCAGCGACTATATATAAAGTGGC TTGGACGCAGGCTGCAAGTTGCGTCCGACACCAGATGAACTGAAGGAGCTCGGGCCAGCGTTCCAACAAAGATGG GATACCTTCTACAAGGACTACCCCGACAAACCGGTCATGCTTGGTGCGAGTATGGCCGCTTGTGGTTCGCCCGCG GGTGTCAAGGAGAAGATGTGGACAGCTGCGTATTACTCCGTGAGCTTTTCTCGTCTACCATGCCCATTTAAGACC GGAACTAAGTGTACCTGTCGTTACCTACTAGGCATACCCATCCTCGACTGGGTGTGTCCATATCGCCGGAGGACG CGACCCATTCGCTCCCTTGGACTACCATCCGGGGTATCTCGACAAGTGAGTTCCAATACCTCGGTCATGCTTTCT GTCTCGTCTGACCATATTCCACGATGACCTCAAAGAGAGGAAGACGTAGCCATGTTCCGATGGGCGTACAAACAC TTGAGGGAGATAGCTCGACGCATGCCGTTCTACCGTGGCGAATACGAGGAGCGTCACCCCAAGTTTCCCGAAGGA AGTGCTGCCGCGTGTAACCTGTCTAGGGAACCCGTCGCAATCGACGCTCCCAAGTTGGTTTACACGAAGGAAGAT GATGAGGCGATCGATAGGTACCATCGGACCACCAGTGCGTTTTGCCACCTACTCCGAGTCTCTCTTGTGACATCG ACTAAATCCGAACAATTCGTGCAGTGGGACCAGCTTGGCATCCGGTAAGTTCCAAGACTTTGAATGTCCTATTAA TGTATCCAATTGACCATAAAATGTAGGCCGGTACTTGTGCGATGAAGCCTCGAGAGCAAGGCGGCGTCCTCGATT CCAGACTCAACGTCTATGGAACCACCAATCTGAAGGTTGCCGGTAAGACGGCTCGCCATGTCTTGATTTCAATCT CCAACTGACGTTCGCCCTTTCTTTTGCATTAGATGTCAGTGTCTGTCCCGAGAACGTCGGAGCCAACACGTACAG TACGGCTTTGGCGGTTGGTCTCAAGGCCACTGTGATCATCGCGGAGGAGCTGGGGATCAAGCTGAGGAACTGAGT GCAGGACATTGGCTGGTAGTCATCCTCTTTTTTTTCAAGTGTGATAAACAATTATGTATACCTCAGCTACTATGT ACTGGTACTCGTTGATGTAAATTAGCCTTAGTCTGAGTGAGCGGACTCACCTCGGATATTTAAGCGCCCATGACT CAACC |
| Length | 2855 |