CopciAB_369187
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_369187 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 369187 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_8:1344961..1348145 | Strand | + |
| Gene length (nt) | 3185 | Transcript length (nt) | 2643 |
| CDS length (nt) | 2559 | Protein length (aa) | 852 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28781 | 34.4 | 4.355E-132 | 443 |
| Auricularia subglabra | Aurde3_1_1203874 | 33.8 | 1.141E-37 | 154 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_369187.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd00882 | Ras_like_GTPase | - | 627 | 713 |
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 242 | 508 |
| Pfam | PF01926 | 50S ribosome-binding GTPase | IPR006073 | 626 | 713 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR006073 | GTP binding domain |
| IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005525 | GTP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
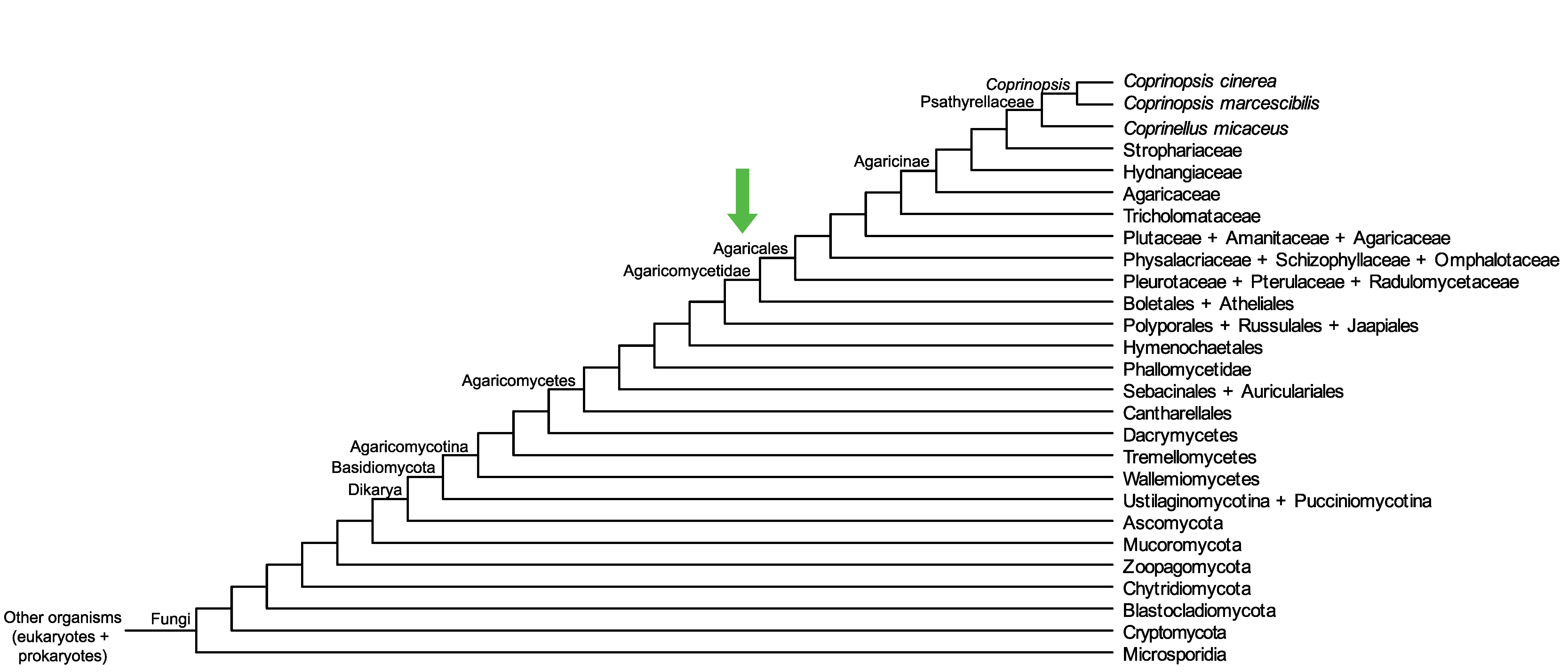
Conservation of CopciAB_369187 across fungi.
Arrow shows the origin of gene family containing CopciAB_369187.
Protein
| Sequence id | CopciAB_369187.T0 |
|---|---|
| Sequence |
>CopciAB_369187.T0 MSSKQTLETLHRRLRECQCGEVSLELLHEKPATWIGHHRDDIAAICAAYSECYPILKDEGISEMLFPRTFILAAV VLGSSEENTSALREILDAGRGLDRVARLAAYRTPKDGLRRILLSETAPTPGFIPRNVVDANFQAPRTSLRPMVNE LLLLETAHIAALVVDCIEQGPGQDTLFLKKDPEVDQNLIDLFQGLLDYPALKSIHPMILRALIHLVKEAALFPRC LVHQDVNIPGQRCTAIGSFGDVWKGDLRGQAVAVKVMRQHVTGIPVDSIIRGYGREAILWSQLHHPNVLPFYGIY LWPSEDSGPTWVCLLSPWMANGNLSQYLEARPALDRMPLVRDIARGLNYLHSLDPSFIHGDIKSNNILVTPSGTA CLADFGLGRLAPDARLGWRSSTVATGGPYLFSAPELLVEDVQPAKGPGSHEVTPAKKKSTQTDVYAFACVVYEMY TGTPRFANLALAEAYRAIISNEPCERPAAMPEPLWAWINRAWDLNPDRRPAMSEFIQEFDPGPPSIQYKWDSKPV SNLRSTTFLAVPPTPIRIQPPRLGTSASSLSASTITSSGASSTTLEVPQCDSPVEDYEVVNGFDDPPPEKSAPRP EKPLPRRPARVPRSQASSFLEDDIIIALMGPTGSGKSTFINCALNDYVAPVSHTITSCTKEVGVYGCFHPNQPGR RVFLVDTPGFESTAMHDRQTLELIAKWLKETYQTHVVLTGVLQFHSIADARVGGTKRDNLDVFRAMCGQDALKNV VYVSTGWEDLDEEEQGERRQHQLVNDYLAEPLQHGCRYQPFLQRDFDAAWAIIDLFQEKRRALKIQEEMVKYRWR LTQTSAYKALSRKFIKVFKFKRGSTMN |
| Length | 852 |
Coding
| Sequence id | CopciAB_369187.T0 |
|---|---|
| Sequence |
>CopciAB_369187.T0 ATGTCAAGCAAACAAACCTTGGAGACATTGCATCGACGGTTACGGGAGTGCCAGTGTGGCGAAGTTAGCCTGGAA CTGTTGCACGAGAAACCAGCGACCTGGATTGGCCACCATCGAGACGACATCGCTGCCATTTGCGCCGCATATTCC GAGTGTTACCCTATTCTGAAGGACGAAGGAATATCCGAAATGTTGTTTCCCCGAACGTTCATTCTGGCGGCAGTC GTCCTGGGAAGTTCCGAGGAAAACACCAGTGCTCTCAGGGAAATCCTCGATGCAGGACGTGGCCTGGATCGAGTG GCCCGCTTGGCCGCGTACAGGACACCCAAGGATGGGTTGCGACGTATTCTACTGTCAGAAACGGCTCCTACGCCT GGATTCATCCCGCGAAATGTCGTAGACGCAAATTTCCAAGCACCGCGTACGTCCCTACGCCCTATGGTCAACGAG CTCTTGCTCTTGGAAACGGCACATATCGCCGCCTTGGTCGTGGATTGCATTGAACAGGGGCCTGGGCAAGACACA TTGTTCTTGAAAAAGGATCCGGAAGTGGACCAAAATCTCATAGATCTATTCCAAGGGCTTCTTGATTACCCTGCC CTCAAGAGTATCCACCCCATGATTTTGCGTGCCCTCATCCACCTCGTGAAGGAAGCTGCACTATTCCCCCGCTGC CTCGTCCATCAGGATGTCAACATCCCAGGCCAGCGCTGTACCGCAATTGGCTCGTTTGGAGATGTTTGGAAGGGG GACCTTCGTGGACAGGCAGTCGCAGTCAAGGTCATGCGACAACATGTAACGGGTATACCGGTGGATTCGATCATC AGAGGATATGGTCGGGAGGCCATCCTGTGGAGCCAGCTTCATCATCCCAATGTCCTGCCATTTTACGGCATTTAC CTCTGGCCAAGCGAAGACTCCGGCCCGACTTGGGTTTGCTTACTTTCTCCTTGGATGGCAAATGGAAACCTTTCC CAGTACCTTGAGGCCAGGCCCGCTTTGGATCGCATGCCCCTGGTTCGAGACATTGCTCGTGGGTTGAACTACCTA CATTCCTTGGACCCCTCCTTCATCCATGGCGATATTAAATCTAATAATATTCTCGTGACCCCCTCTGGGACTGCG TGCTTGGCAGACTTCGGACTAGGCCGTCTCGCACCGGATGCCCGCCTGGGCTGGCGTTCATCGACTGTTGCAACG GGAGGGCCGTATCTCTTTTCGGCCCCGGAATTGCTTGTCGAGGATGTGCAACCTGCCAAAGGACCAGGTTCTCAC GAGGTGACACCAGCGAAAAAGAAGTCAACCCAGACGGACGTCTACGCATTTGCCTGTGTTGTTTACGAGATGTAC ACGGGCACACCTAGATTTGCAAACCTTGCGTTGGCTGAAGCCTACCGAGCAATCATTTCGAATGAGCCTTGCGAG CGACCTGCTGCGATGCCAGAACCACTATGGGCTTGGATTAATCGGGCATGGGACCTCAACCCCGACCGAAGACCA GCGATGTCTGAATTCATCCAAGAGTTCGATCCTGGGCCACCATCGATACAGTACAAGTGGGACTCCAAGCCCGTC TCGAACCTGCGATCCACGACATTTTTGGCGGTGCCTCCTACTCCGATTCGCATTCAGCCACCCAGGTTGGGGACA AGTGCGTCCTCACTCAGTGCGAGCACCATAACTAGTTCGGGTGCCTCTTCTACGACCCTCGAGGTACCGCAATGC GATAGTCCGGTTGAAGACTATGAAGTTGTCAATGGCTTTGACGACCCTCCGCCAGAGAAGTCCGCCCCAAGGCCC GAGAAGCCTCTCCCAAGACGTCCGGCCAGAGTGCCCAGGTCACAGGCGTCGTCATTCCTGGAGGATGACATTATA ATTGCGTTAATGGGCCCGACCGGTAGTGGGAAAAGCACATTCATTAATTGCGCGTTGAACGACTATGTTGCGCCC GTCTCGCACACGATAACATCCTGTACGAAGGAAGTTGGAGTGTACGGGTGCTTCCATCCGAACCAGCCGGGTCGG CGAGTGTTCCTCGTCGACACTCCAGGGTTCGAAAGTACAGCCATGCACGATCGCCAAACTTTGGAGTTGATCGCT AAATGGCTAAAGGAAACGTATCAAACGCACGTTGTGCTCACAGGGGTTTTGCAATTCCACTCGATCGCTGATGCC AGGGTTGGAGGCACAAAAAGAGACAACTTAGATGTCTTCCGAGCGATGTGTGGCCAGGATGCGCTCAAGAACGTT GTCTACGTCTCGACAGGTTGGGAGGACTTGGATGAAGAAGAGCAAGGTGAACGAAGGCAGCATCAGCTTGTCAAT GATTACCTTGCAGAACCTCTGCAGCATGGGTGTCGCTACCAGCCTTTCCTCCAGCGAGACTTCGACGCGGCTTGG GCTATCATCGACCTTTTCCAAGAGAAAAGAAGGGCGCTGAAAATACAGGAAGAGATGGTGAAATACAGATGGCGT CTAACGCAAACATCTGCCTATAAAGCTCTATCTCGCAAATTTATTAAGGTGTTTAAATTCAAACGAGGTAGTACC ATGAACTGA |
| Length | 2559 |
Transcript
| Sequence id | CopciAB_369187.T0 |
|---|---|
| Sequence |
>CopciAB_369187.T0 ACCCTATACCTTAACCCTTCACCGTTAGTTCTCGGTATCGATGTCAAGCAAACAAACCTTGGAGACATTGCATCG ACGGTTACGGGAGTGCCAGTGTGGCGAAGTTAGCCTGGAACTGTTGCACGAGAAACCAGCGACCTGGATTGGCCA CCATCGAGACGACATCGCTGCCATTTGCGCCGCATATTCCGAGTGTTACCCTATTCTGAAGGACGAAGGAATATC CGAAATGTTGTTTCCCCGAACGTTCATTCTGGCGGCAGTCGTCCTGGGAAGTTCCGAGGAAAACACCAGTGCTCT CAGGGAAATCCTCGATGCAGGACGTGGCCTGGATCGAGTGGCCCGCTTGGCCGCGTACAGGACACCCAAGGATGG GTTGCGACGTATTCTACTGTCAGAAACGGCTCCTACGCCTGGATTCATCCCGCGAAATGTCGTAGACGCAAATTT CCAAGCACCGCGTACGTCCCTACGCCCTATGGTCAACGAGCTCTTGCTCTTGGAAACGGCACATATCGCCGCCTT GGTCGTGGATTGCATTGAACAGGGGCCTGGGCAAGACACATTGTTCTTGAAAAAGGATCCGGAAGTGGACCAAAA TCTCATAGATCTATTCCAAGGGCTTCTTGATTACCCTGCCCTCAAGAGTATCCACCCCATGATTTTGCGTGCCCT CATCCACCTCGTGAAGGAAGCTGCACTATTCCCCCGCTGCCTCGTCCATCAGGATGTCAACATCCCAGGCCAGCG CTGTACCGCAATTGGCTCGTTTGGAGATGTTTGGAAGGGGGACCTTCGTGGACAGGCAGTCGCAGTCAAGGTCAT GCGACAACATGTAACGGGTATACCGGTGGATTCGATCATCAGAGGATATGGTCGGGAGGCCATCCTGTGGAGCCA GCTTCATCATCCCAATGTCCTGCCATTTTACGGCATTTACCTCTGGCCAAGCGAAGACTCCGGCCCGACTTGGGT TTGCTTACTTTCTCCTTGGATGGCAAATGGAAACCTTTCCCAGTACCTTGAGGCCAGGCCCGCTTTGGATCGCAT GCCCCTGGTTCGAGACATTGCTCGTGGGTTGAACTACCTACATTCCTTGGACCCCTCCTTCATCCATGGCGATAT TAAATCTAATAATATTCTCGTGACCCCCTCTGGGACTGCGTGCTTGGCAGACTTCGGACTAGGCCGTCTCGCACC GGATGCCCGCCTGGGCTGGCGTTCATCGACTGTTGCAACGGGAGGGCCGTATCTCTTTTCGGCCCCGGAATTGCT TGTCGAGGATGTGCAACCTGCCAAAGGACCAGGTTCTCACGAGGTGACACCAGCGAAAAAGAAGTCAACCCAGAC GGACGTCTACGCATTTGCCTGTGTTGTTTACGAGATGTACACGGGCACACCTAGATTTGCAAACCTTGCGTTGGC TGAAGCCTACCGAGCAATCATTTCGAATGAGCCTTGCGAGCGACCTGCTGCGATGCCAGAACCACTATGGGCTTG GATTAATCGGGCATGGGACCTCAACCCCGACCGAAGACCAGCGATGTCTGAATTCATCCAAGAGTTCGATCCTGG GCCACCATCGATACAGTACAAGTGGGACTCCAAGCCCGTCTCGAACCTGCGATCCACGACATTTTTGGCGGTGCC TCCTACTCCGATTCGCATTCAGCCACCCAGGTTGGGGACAAGTGCGTCCTCACTCAGTGCGAGCACCATAACTAG TTCGGGTGCCTCTTCTACGACCCTCGAGGTACCGCAATGCGATAGTCCGGTTGAAGACTATGAAGTTGTCAATGG CTTTGACGACCCTCCGCCAGAGAAGTCCGCCCCAAGGCCCGAGAAGCCTCTCCCAAGACGTCCGGCCAGAGTGCC CAGGTCACAGGCGTCGTCATTCCTGGAGGATGACATTATAATTGCGTTAATGGGCCCGACCGGTAGTGGGAAAAG CACATTCATTAATTGCGCGTTGAACGACTATGTTGCGCCCGTCTCGCACACGATAACATCCTGTACGAAGGAAGT TGGAGTGTACGGGTGCTTCCATCCGAACCAGCCGGGTCGGCGAGTGTTCCTCGTCGACACTCCAGGGTTCGAAAG TACAGCCATGCACGATCGCCAAACTTTGGAGTTGATCGCTAAATGGCTAAAGGAAACGTATCAAACGCACGTTGT GCTCACAGGGGTTTTGCAATTCCACTCGATCGCTGATGCCAGGGTTGGAGGCACAAAAAGAGACAACTTAGATGT CTTCCGAGCGATGTGTGGCCAGGATGCGCTCAAGAACGTTGTCTACGTCTCGACAGGTTGGGAGGACTTGGATGA AGAAGAGCAAGGTGAACGAAGGCAGCATCAGCTTGTCAATGATTACCTTGCAGAACCTCTGCAGCATGGGTGTCG CTACCAGCCTTTCCTCCAGCGAGACTTCGACGCGGCTTGGGCTATCATCGACCTTTTCCAAGAGAAAAGAAGGGC GCTGAAAATACAGGAAGAGATGGTGAAATACAGATGGCGTCTAACGCAAACATCTGCCTATAAAGCTCTATCTCG CAAATTTATTAAGGTGTTTAAATTCAAACGAGGTAGTACCATGAACTGATGCGTGTATGTCATATCAATATAAAA GTCTATGCATATCGGCAC |
| Length | 2643 |
Gene
| Sequence id | CopciAB_369187.T0 |
|---|---|
| Sequence |
>CopciAB_369187.T0 ACCCTATACCTTAACCCTTCACCGTTAGTTCTCGGTATCGATGTCAAGCAAACAAACCTTGGAGACATTGCATCG ACGGTTACGGGAGTGCCAGTGTGGCGAAGTTAGCCTGGAACTGTTGCACGAGAAACCAGCGACCTGGATTGGCCA CCATCGAGACGACATCGCTGCCATTTGCGCCGCATATTCCGAGTGTTACCCTATTCTGAAGGACGAAGGAATATC CGAAATGTTGTTTCCCGTTCGTAAAGCTTTTCATTCCACATCCATGCATCCGGCTGACTGGGACTGACGATCGAT AGCGAACGTTCATTCTGGCGGCAGTCGTCCTGGGAAGTTCCGAGGAAAACACCAGGTCGGTTGCTTCGCGCCTCA AAGTTTCCTCTTTCGCTCATCGACGATCCTATTTGGTCTTCAGTGCTCTCAGGGAAATCCTCGATGCAGGACGTG GCCTGGATCGAGTGGCCCGCTTGGCCGCGTACAGGACACCCAAGGATGGGTTGCGACGTATTCTACTGTCAGAAA CGGCTCCTACGCCTGGATTCATCCCGCGAAATGTCGTAGACGCAAATTTCCAAGCACCGCGTACGTCCCTACGCC CTATGGTCAACGAGCTCTTGCTCTTGGAAACGGCACATATCGCCGCCTTGGTCGTGGATTGCATTGAACAGGGGC CTGGGCAAGACACATTGTTCTTGAAAAAGGATCCGGAAGTGGACCAAAATCTCATAGATCTATTCCAAGGGGTGC GTCCGTTTACTTCTCACCTTCTAGTAAAGCGTTACTCAATGTGCAACGTCGACCGTAGCTTCTTGATTACCCTGC CCTCAAGAGTATCCACCCCATGATTTTGCGTGCCCTCATCCACCTCGTGAAGGAAGCTGCACTATTCCCCCGCTG CCTCGTCCATCAGGATGTCAACATCCCAGGCCAGCGCTGTACCGCAATTGGCTCGTTTGGAGATGTTTGGAAGGG GGACCTTCGTGGACAGGCAGTCGCAGTCAAGGTCATGCGACAACATGTAACGGGTATACCGGTGGATTCGATCAT CAGAGTGAGTAACGTCGACGTCTGTGCAGGGATTCTCTTATCATTCTTAAACAGGGATATGGTCGGGAGGCCATC CTGTGGAGCCAGCTTCATCATCCCAATGTCCTGCCATTTTACGGCATTTACCTCTGGCCAAGCGAAGACTCCGGC CCGACTTGGGTTTGCTTACTTTCTCCTTGGATGGCAAATGGAAACCTTTCCCAGTACCTTGAGGCCAGGCCCGCT TTGGATCGCATGCCCCTGGTGAGCTTAAGACCGCACGGCGTGTCAACTTGTCGCTCATCTTCTGTCAGGTTCGAG ACATTGCTCGTGGGTTGAACTACCTACATTCCTTGGACCCCTCCTTCATCCATGGCGATATTAAATCTGTAAGAT GCACAGCCCACGTTCAGTAACACTCTTTAACAATGCTCTTAGAATAATATTCTCGTGACCCCCTCTGGGACTGCG TGCTTGGCAGACTTCGGACTAGGCCGTCTCGCACCGGATGCCCGCCTGGGCTGGCGTTCATCGACTGTTGCAACG GGAGGGCCGTATCTCTTTTCGGCCCCGGAATTGCTTGTCGAGGATGTGCAACCTGCCAAAGGACCAGGTTCTCAC GAGGTGACACCAGCGAAAAAGAAGTCAACCCAGACGGACGTCTACGCATTTGCCTGTGTTGTTTACGAGGTACGG TGGTTCACCTCTTACCGGCTCCGGTTGCTCAACTTATCCAGATGTACACGGGCACACCTAGATTTGCAAACCTTG CGTTGGCTGAAGCCTACCGAGCAATCATTTCGAATGAGCCTTGCGAGCGACCTGCTGCGATGCCAGAACCACTAT GGGCTTGGATTAATCGGGCATGGGACCTCAACCCCGACCGAAGACCAGCGATGTCTGAATTCATCCAAGAGTTCG ATCCTGGGCCACCATCGATACAGTACAAGTGGGACTCCAAGCCCGTCTCGAACCTGCGATCCACGACATTTTTGG CGGTGCCTCCTACTCCGATTCGCATTCAGCCACCCAGGTTGGGGACAAGTGCGTCCTCACTCAGTGCGAGCACCA TAACTAGTTCGGGTGCCTCTTCTACGACCCTCGAGGTACCGCAATGCGATAGTCCGGTTGAAGACTATGAAGTTG TCAATGGCTTTGACGACCCTCCGCCAGAGAAGTCCGCCCCAAGGCCCGAGAAGCCTCTCCCAAGACGTCCGGCCA GAGTGCCCAGGTCACAGGCGTCGTCATTCCTGGAGGATGACATTATAATTGCGTTGGTATTCTTTCCCACTAGTG TCGAATTGCTGGGCTCACCCACCACTTCCAGGTTAATGGGCCCGACCGGTAGTGGGAAAAGCACAGTGAGTGGTT TACCGACAGGCACCCTTCCTCATTGACACTGATGCATTTTGAGCAGTTCATTAATTGCGCGTTGAACGACTATGT TGCGCCCGTCTCGCACACGATAACATCCTGTACGAAGGAAGTTGGAGTGTACGGGTGCTTCCATCCGAACCAGCC GGGTCGGCGAGTGTTCCTCGTCGACACTCCAGGGTTCGAAAGTACAGCCATGCACGATCGCCAAACTTTGGAGTT GATCGCTAAATGGCTAAAGGAAACGTGAGCTTTTTTCCGGACGCATTCCCGACGCCCAACCTAAATTGTTCCAGG TATCAAACGCACGTTGTGCTCACAGGGGTTTTGCAATTCCACTCGATCGCTGATGCCAGGGTTGGAGGCACAAAA AGAGACAACTTAGATGTCTTCCGAGCGATGTGTGGCCAGGATGCGCTCAAGAACGTTGTCTACGTCTCGACAGGT TGGGAGGACTTGGATGAAGAAGAGCAAGGTGAACGAAGGCAGCATCAGCTTGTCAATGATTACCTTGCAGAACCT CTGCAGCATGGGTGTCGCTACCAGCCTTTCCTCCAGCGAGACTTCGACGCGGCTTGGGCTATCATCGACCTTTTC CAAGAGAAAAGAAGGGCGCTGAAAATACAGGAAGAGATGGTGAAATACAGATGGCGTCTAACGCAAACATCTGCC TATAAAGCTCTATCTCGCAAATTTATTAAGGTGTTTAAATTCAAACGAGGTAGTACCATGAACTGATGCGTGTAT GTCATATCAATATAAAAGTCTATGCATATCGGCAC |
| Length | 3185 |