CopciAB_370595
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_370595 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | SNT2 | Synonyms | 370595 |
| Uniprot id | Functional description | PHD-zinc-finger like domain | |
| Location | scaffold_6:1961252..1965553 | Strand | - |
| Gene length (nt) | 4302 | Transcript length (nt) | 3463 |
| CDS length (nt) | 2817 | Protein length (aa) | 938 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | NCU07412 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB15348 | 72.4 | 0 | 1498 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_54474 | 69.9 | 0 | 1428 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_205606 | 69.9 | 0 | 1427 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1105423 | 65.5 | 0 | 1302 |
| Pleurotus ostreatus PC9 | PleosPC9_1_129613 | 65.3 | 0 | 1300 |
| Agrocybe aegerita | Agrae_CAA7265141 | 63.6 | 0 | 1246 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1560541 | 64.5 | 0 | 1222 |
| Flammulina velutipes | Flave_chr06AA00938 | 61.5 | 0 | 1183 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_50126 | 58.9 | 0 | 1134 |
| Lentinula edodes NBRC 111202 | Lenedo1_1259524 | 57.8 | 0 | 1124 |
| Schizophyllum commune H4-8 | Schco3_2619667 | 55.6 | 0 | 1096 |
| Grifola frondosa | Grifr_OBZ73631 | 57.5 | 1.482E-291 | 1005 |
| Auricularia subglabra | Aurde3_1_1021623 | 48.5 | 1.498E-301 | 942 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_18564 | 55.9 | 5.553E-288 | 902 |
| Lentinula edodes B17 | Lened_B_1_1_7625 | 57 | 1.754E-106 | 369 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | SNT2 |
|---|---|
| Protein id | CopciAB_370595.T0 |
| Description | PHD-zinc-finger like domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd04710 | BAH_fungalPHD | - | 7 | 142 |
| CDD | cd15497 | PHD1_Snt2p_like | - | 184 | 230 |
| CDD | cd15571 | ePHD | - | 676 | 788 |
| Pfam | PF01426 | BAH domain | IPR001025 | 18 | 144 |
| Pfam | PF00628 | PHD-finger | IPR019787 | 184 | 232 |
| Pfam | PF13832 | PHD-zinc-finger like domain | - | 701 | 786 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR013088 | Zinc finger, NHR/GATA-type |
| IPR019787 | Zinc finger, PHD-finger |
| IPR001965 | Zinc finger, PHD-type |
| IPR011011 | Zinc finger, FYVE/PHD-type |
| IPR013083 | Zinc finger, RING/FYVE/PHD-type |
| IPR029617 | Lid2 complex component Snt2 |
| IPR001025 | Bromo adjacent homology (BAH) domain |
| IPR043151 | Bromo adjacent homology (BAH) domain superfamily |
| IPR000679 | Zinc finger, GATA-type |
| IPR009057 | Homeobox-like domain superfamily |
| IPR034732 | Extended PHD (ePHD) domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0006355 | regulation of transcription, DNA-templated | BP |
| GO:0008270 | zinc ion binding | MF |
| GO:0003682 | chromatin binding | MF |
| GO:0043565 | sequence-specific DNA binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | PHD-zinc-finger like domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| zinc finger, gata-type |
| helix-turn-helix |
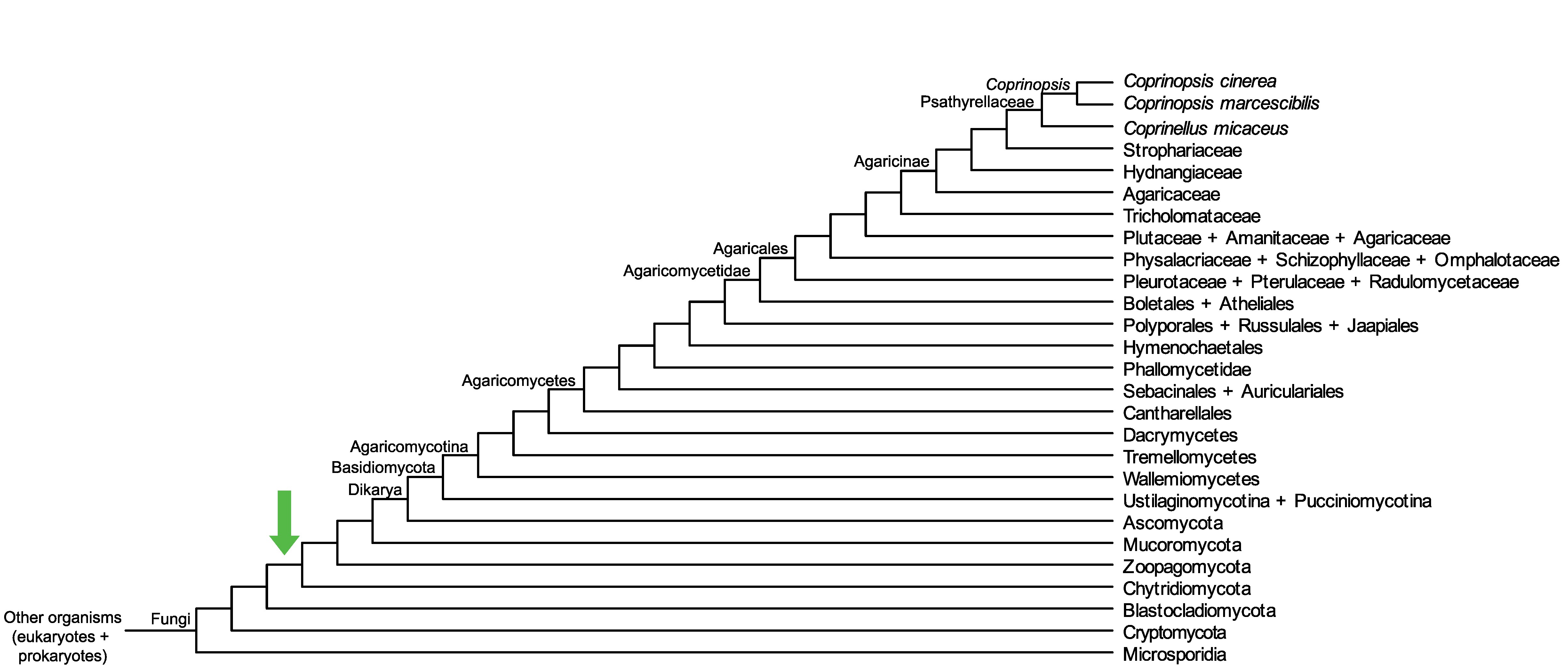
Conservation of CopciAB_370595 across fungi.
Arrow shows the origin of gene family containing CopciAB_370595.
Protein
| Sequence id | CopciAB_370595.T0 |
|---|---|
| Sequence |
>CopciAB_370595.T0 MNMNDDEIPVYLKNGDLVKVNDHVYCSPSWDTRDGTPYSVGRIMQFLPPADAPKGEEGKNYHYTRVRLAWYYRPS DVSDRPVADSRLLLAAIYSEVCDINQLRAKCHVVHRDKISDLSGWKKRPDRFYFNRLFDPYIKKEFEVIPSRDVR NLPDHIREVLISRYEYIVAEKEVIPDLTDHIRLCDNCQKWCPSADTVQCDRCKRFFHMRCVQPPLLAKPTRGYGW TCAPCSRRHEEEVDSHDVRPSSSAAGKAHRSNAPPARGRGRPRKDKTLAEKEENLPVKHFKMWPFRYFGQHTVAE DTLDEEDLIFPRTATRHGPKYQANVPPGPDPYNYPPDLEERGGDNTVEVLGVINSLTESEVAEVEACKKGLTNDP KLRYSVDWLTEVIFRFSDAALAARPLTAVNMKNPNRIQKWKKYETPYTDRPWSHDEVVAFEDAVALHGPELRAAK DEVPNRSMPEVVRFFGHWKNSKLKEENKRLKTSGPPAKPIHRQYRSAAERSDGQRVGLGDDEGSIVAQPSKSPSC GACRTRESPQWWKAPKGLPTNILCETCGTNWRKYADLNVRPIREESLPTSKAKTSEKREGTPPAGPSAKRARTSA SVQSTPPPVVSAAPQVKCFACHKQGPLGKVLRCKKCQFRAHAGCVGAVVGPAKVDEWTCELCENEETMEASINYD CLLCPRSGPEEKKQKPWPPADTFLRACKPTEGQGWTHVICAVFLPELSFTDASRLRLVEGMSTIAKHRWTTKCCL CGEMEGAVVKCSDCYREFHVSCAWKEGHKFGFEIQPVKSSRRDTTITTTFKGESGCMSALISCKMHDHNKRDIYD ICETNEGGETALQVYCQAYKQAQVGQAHGLLRKAKRLDALLNLRDPHAHGMQPVQELPTTEPECSKCNTQFSPAF YPVVDLSPTASKETWLCHRCHFEANVEVNGKAMGMMIS |
| Length | 938 |
Coding
| Sequence id | CopciAB_370595.T0 |
|---|---|
| Sequence |
>CopciAB_370595.T0 ATGAACATGAACGACGACGAGATTCCAGTTTACCTGAAAAACGGCGATCTCGTCAAAGTTAATGACCACGTTTAC TGTTCCCCCAGTTGGGACACTCGGGATGGCACGCCCTACTCAGTCGGACGGATAATGCAATTCTTGCCACCCGCA GATGCACCCAAGGGTGAGGAGGGCAAAAATTACCATTATACGAGGGTCCGTCTCGCCTGGTATTATCGTCCCAGC GACGTCAGCGACCGCCCTGTAGCGGATTCACGTTTGCTTTTGGCGGCCATATACTCGGAAGTGTGCGACATCAAT CAGCTTCGCGCCAAGTGCCACGTGGTACATCGAGACAAAATATCCGATTTATCTGGCTGGAAGAAACGTCCTGAC CGGTTCTACTTCAACCGCCTTTTCGACCCCTACATCAAGAAAGAGTTTGAAGTTATCCCATCGAGGGACGTACGA AACCTCCCCGACCACATTCGAGAGGTTCTTATTTCACGATATGAATATATCGTCGCGGAGAAAGAGGTCATCCCT GATTTGACCGACCACATCCGCCTTTGTGACAACTGTCAGAAATGGTGCCCCAGCGCTGATACAGTCCAATGTGAT CGCTGTAAACGCTTCTTCCATATGCGCTGCGTCCAGCCTCCGCTTCTCGCGAAACCTACCCGTGGGTATGGCTGG ACCTGTGCCCCCTGTTCTCGACGGCACGAAGAAGAGGTAGACAGCCATGACGTCCGACCTTCCAGCTCGGCCGCG GGGAAGGCCCATAGGTCGAATGCCCCACCTGCTCGAGGACGAGGTCGGCCCAGGAAGGACAAGACCCTGGCTGAG AAGGAGGAGAATCTTCCTGTGAAACATTTCAAGATGTGGCCTTTCAGATATTTCGGTCAACATACTGTGGCGGAA GATACATTGGACGAAGAAGATCTAATATTCCCTCGTACAGCCACAAGGCACGGTCCAAAATACCAGGCAAATGTC CCTCCAGGACCGGATCCATACAATTACCCCCCAGACCTCGAAGAGCGTGGTGGTGACAACACAGTGGAAGTTCTC GGCGTTATCAACTCATTGACAGAGTCGGAAGTGGCTGAAGTGGAAGCATGCAAGAAGGGCCTAACCAATGACCCC AAGCTCCGTTACAGCGTTGACTGGTTAACAGAAGTCATCTTCCGTTTCTCTGATGCAGCTCTCGCGGCGCGTCCA TTAACGGCTGTCAACATGAAGAATCCCAATCGAATACAGAAATGGAAGAAATACGAAACGCCGTACACAGATCGT CCCTGGTCACACGACGAAGTCGTCGCCTTTGAAGATGCAGTCGCCCTTCACGGTCCCGAGCTTCGTGCTGCCAAG GACGAAGTGCCAAACCGCTCGATGCCAGAGGTCGTTCGTTTCTTCGGGCACTGGAAGAACTCCAAGCTCAAGGAG GAAAACAAACGTCTCAAAACCTCCGGCCCACCTGCCAAACCAATACATCGCCAATACCGATCCGCTGCGGAACGA AGCGATGGTCAACGCGTCGGCTTAGGTGACGATGAAGGTTCGATCGTCGCACAACCGTCCAAGTCCCCAAGTTGT GGTGCTTGTCGTACACGTGAATCGCCTCAATGGTGGAAAGCCCCCAAGGGATTGCCAACGAACATCCTTTGCGAG ACATGCGGTACGAACTGGAGGAAGTATGCTGATCTCAATGTCCGTCCAATACGAGAAGAGTCCCTGCCGACTTCG AAGGCGAAGACCTCAGAGAAGCGTGAGGGAACACCCCCGGCTGGTCCTAGCGCCAAGCGGGCAAGAACTAGCGCT TCTGTGCAGTCAACCCCACCACCAGTGGTGTCTGCAGCCCCGCAGGTCAAATGCTTCGCTTGCCACAAACAGGGT CCGCTCGGCAAGGTCCTTCGATGCAAAAAGTGCCAATTCCGGGCTCATGCAGGATGTGTGGGCGCAGTCGTCGGT CCAGCCAAGGTGGACGAGTGGACGTGCGAACTGTGCGAGAACGAAGAGACCATGGAAGCTTCCATTAACTACGAC TGCCTCCTTTGCCCTCGTTCTGGACCCGAAGAGAAGAAGCAGAAGCCATGGCCACCAGCCGATACCTTCTTACGA GCATGCAAACCAACAGAAGGACAGGGCTGGACTCATGTCATCTGTGCAGTGTTTTTACCGGAGCTGAGCTTTACG GACGCGTCTCGTTTACGTCTTGTAGAGGGGATGAGTACCATTGCGAAACATCGTTGGACAACGAAATGTTGCCTG TGTGGCGAGATGGAGGGAGCGGTAGTGAAATGCAGCGACTGCTATCGAGAATTCCATGTATCTTGCGCCTGGAAA GAGGGGCACAAGTTCGGCTTCGAGATCCAACCAGTGAAGAGCAGCCGAAGAGATACGACCATCACAACGACGTTC AAGGGTGAATCAGGCTGCATGAGTGCCCTCATCTCTTGTAAGATGCATGATCACAATAAACGTGACATCTATGAC ATTTGCGAAACCAACGAGGGTGGCGAGACCGCGCTGCAGGTGTATTGCCAGGCGTACAAGCAAGCGCAGGTAGGC CAAGCCCATGGCTTGCTGCGCAAAGCCAAACGGCTGGACGCCCTGCTCAACTTACGCGATCCCCACGCACACGGG ATGCAACCGGTACAAGAACTCCCCACCACGGAACCCGAATGCAGCAAGTGCAACACGCAATTCTCTCCGGCTTTC TATCCCGTCGTCGATTTGTCGCCTACAGCTTCCAAAGAGACGTGGCTTTGCCATCGATGTCACTTTGAGGCCAAC GTCGAGGTCAACGGCAAGGCAATGGGTATGATGATATCATGA |
| Length | 2817 |
Transcript
| Sequence id | CopciAB_370595.T0 |
|---|---|
| Sequence |
>CopciAB_370595.T0 ACCTCTTGTTTCCTCTCCTCCACCTTCTCCTTATTTCTGACGCCCAAAGCCTGAATTCCCTTGTTTTACGGGCTC TAAATATCTGAGGGTGTTTTATACTCGTTCACTCGACCCATTTCTTCACCTTCTTTTCTATTTACCCCATTTTCC CTTTTTTGGAGCTCAGATTTCGAGCTCAGCCGCGTAACCACCCATCATGAACATGAACGACGACGAGATTCCAGT TTACCTGAAAAACGGCGATCTCGTCAAAGTTAATGACCACGTTTACTGTTCCCCCAGTTGGGACACTCGGGATGG CACGCCCTACTCAGTCGGACGGATAATGCAATTCTTGCCACCCGCAGATGCACCCAAGGGTGAGGAGGGCAAAAA TTACCATTATACGAGGGTCCGTCTCGCCTGGTATTATCGTCCCAGCGACGTCAGCGACCGCCCTGTAGCGGATTC ACGTTTGCTTTTGGCGGCCATATACTCGGAAGTGTGCGACATCAATCAGCTTCGCGCCAAGTGCCACGTGGTACA TCGAGACAAAATATCCGATTTATCTGGCTGGAAGAAACGTCCTGACCGGTTCTACTTCAACCGCCTTTTCGACCC CTACATCAAGAAAGAGTTTGAAGTTATCCCATCGAGGGACGTACGAAACCTCCCCGACCACATTCGAGAGGTTCT TATTTCACGATATGAATATATCGTCGCGGAGAAAGAGGTCATCCCTGATTTGACCGACCACATCCGCCTTTGTGA CAACTGTCAGAAATGGTGCCCCAGCGCTGATACAGTCCAATGTGATCGCTGTAAACGCTTCTTCCATATGCGCTG CGTCCAGCCTCCGCTTCTCGCGAAACCTACCCGTGGGTATGGCTGGACCTGTGCCCCCTGTTCTCGACGGCACGA AGAAGAGGTAGACAGCCATGACGTCCGACCTTCCAGCTCGGCCGCGGGGAAGGCCCATAGGTCGAATGCCCCACC TGCTCGAGGACGAGGTCGGCCCAGGAAGGACAAGACCCTGGCTGAGAAGGAGGAGAATCTTCCTGTGAAACATTT CAAGATGTGGCCTTTCAGATATTTCGGTCAACATACTGTGGCGGAAGATACATTGGACGAAGAAGATCTAATATT CCCTCGTACAGCCACAAGGCACGGTCCAAAATACCAGGCAAATGTCCCTCCAGGACCGGATCCATACAATTACCC CCCAGACCTCGAAGAGCGTGGTGGTGACAACACAGTGGAAGTTCTCGGCGTTATCAACTCATTGACAGAGTCGGA AGTGGCTGAAGTGGAAGCATGCAAGAAGGGCCTAACCAATGACCCCAAGCTCCGTTACAGCGTTGACTGGTTAAC AGAAGTCATCTTCCGTTTCTCTGATGCAGCTCTCGCGGCGCGTCCATTAACGGCTGTCAACATGAAGAATCCCAA TCGAATACAGAAATGGAAGAAATACGAAACGCCGTACACAGATCGTCCCTGGTCACACGACGAAGTCGTCGCCTT TGAAGATGCAGTCGCCCTTCACGGTCCCGAGCTTCGTGCTGCCAAGGACGAAGTGCCAAACCGCTCGATGCCAGA GGTCGTTCGTTTCTTCGGGCACTGGAAGAACTCCAAGCTCAAGGAGGAAAACAAACGTCTCAAAACCTCCGGCCC ACCTGCCAAACCAATACATCGCCAATACCGATCCGCTGCGGAACGAAGCGATGGTCAACGCGTCGGCTTAGGTGA CGATGAAGGTTCGATCGTCGCACAACCGTCCAAGTCCCCAAGTTGTGGTGCTTGTCGTACACGTGAATCGCCTCA ATGGTGGAAAGCCCCCAAGGGATTGCCAACGAACATCCTTTGCGAGACATGCGGTACGAACTGGAGGAAGTATGC TGATCTCAATGTCCGTCCAATACGAGAAGAGTCCCTGCCGACTTCGAAGGCGAAGACCTCAGAGAAGCGTGAGGG AACACCCCCGGCTGGTCCTAGCGCCAAGCGGGCAAGAACTAGCGCTTCTGTGCAGTCAACCCCACCACCAGTGGT GTCTGCAGCCCCGCAGGTCAAATGCTTCGCTTGCCACAAACAGGGTCCGCTCGGCAAGGTCCTTCGATGCAAAAA GTGCCAATTCCGGGCTCATGCAGGATGTGTGGGCGCAGTCGTCGGTCCAGCCAAGGTGGACGAGTGGACGTGCGA ACTGTGCGAGAACGAAGAGACCATGGAAGCTTCCATTAACTACGACTGCCTCCTTTGCCCTCGTTCTGGACCCGA AGAGAAGAAGCAGAAGCCATGGCCACCAGCCGATACCTTCTTACGAGCATGCAAACCAACAGAAGGACAGGGCTG GACTCATGTCATCTGTGCAGTGTTTTTACCGGAGCTGAGCTTTACGGACGCGTCTCGTTTACGTCTTGTAGAGGG GATGAGTACCATTGCGAAACATCGTTGGACAACGAAATGTTGCCTGTGTGGCGAGATGGAGGGAGCGGTAGTGAA ATGCAGCGACTGCTATCGAGAATTCCATGTATCTTGCGCCTGGAAAGAGGGGCACAAGTTCGGCTTCGAGATCCA ACCAGTGAAGAGCAGCCGAAGAGATACGACCATCACAACGACGTTCAAGGGTGAATCAGGCTGCATGAGTGCCCT CATCTCTTGTAAGATGCATGATCACAATAAACGTGACATCTATGACATTTGCGAAACCAACGAGGGTGGCGAGAC CGCGCTGCAGGTGTATTGCCAGGCGTACAAGCAAGCGCAGGTAGGCCAAGCCCATGGCTTGCTGCGCAAAGCCAA ACGGCTGGACGCCCTGCTCAACTTACGCGATCCCCACGCACACGGGATGCAACCGGTACAAGAACTCCCCACCAC GGAACCCGAATGCAGCAAGTGCAACACGCAATTCTCTCCGGCTTTCTATCCCGTCGTCGATTTGTCGCCTACAGC TTCCAAAGAGACGTGGCTTTGCCATCGATGTCACTTTGAGGCCAACGTCGAGGTCAACGGCAAGGCAATGGGTAT GATGATATCATGAAGCGACGAGGCATGAAGCGACGAGGCACGATGCTCTTTCTTATCAATGAACGCGACACCACT GGCCTCTCCACACGCGTAGTCGTCCAAGGAGTCTGAATGGGCGAATGCAACATGTGAATGCCAAGGAGAAAGGAA GAAGCGAGAGACGAAAGACGAAAGACAACTTGACTATGATGATACAATAAACTACAGCAGGGGATGGGGTGTCGA TTGAAATACATCCGGGTGTCTGTCTGTTCTGAAAGAGCGAACGAATCACGAATAACCATAGGAAGGGGGTAGTAG GTAGTAGTAATACGTATGTAATAACGCAAAGCAGGTCCTTACGCGCGAACTCTCTTGTAGAAAAGGACATACGCC TTTTGGTTCTGTAGTTTTCGTTGTTAGTATTATTAGCACGCTGTCTTTTTAATTTATTGACGACCAATAGGAGTA TGACTTACCACGA |
| Length | 3463 |
Gene
| Sequence id | CopciAB_370595.T0 |
|---|---|
| Sequence |
>CopciAB_370595.T0 ACCTCTTGTTTCCTCTCCTCCACCTTCTCCTTATTTCTGACGCCCAAAGCCTGAATTCCCTTGTTTTACGGGCTC TAAATATCTGAGGGTGTTTTATACTCGTTCACTCGACCCATTTCTTCACCTTCTTTTCTATTTACCCCATTTTCC CTTTTTTGGAGCTCAGATTTCGAGCTCAGCCGCGTAACCACCCATCATGAACATGAACGACGACGAGATTCCAGT TTACCTGAAAAACGGCGATCTCGTCAAAGTTAATGGTAATATTCTCAATTCAAAGTATCCCCCAGGTAGTACACA ACCCTTACATATTTTTCAGACCACGTTTACTGTTCCCCCAGTTGGGACACTCGGGATGGCACGCCCTACTCAGTC GGACGGATAATGCAATTCTTGCCACCCGCAGATGCACCCAAGGGTGAGGAGGGCAAAAATTACCATTATACGAGG GTCCGTCTCGCCTGGTATTATCGTCCCAGCGACGTCAGCGACCGCCCTGTAGCGGATTCACGTTTGCTTTTGGCG GCCATATACTCGGAAGTGTGCGACATCAATCAGCTTCGCGCCAAGTGCCACGTGGTACATCGAGACAAAATATCC GATTTATCTGGCTGGAAGAAACGTCCTGACCGGTTCTACTTCAACCGCCTTTTCGACCCCTACATCAAGAAAGAG TTTGAAGTTATCCCATCGAGGGACGTACGAAACCGTGAGTAAATCAGTCTTTGTCATAAATATTGATCATTTTCC TAATTTAAACCTTCCTTCTCCATCTAGTCCCCGACCACATTCGAGAGGTTCTTATTTCACGATATGAATATATCG TCGCGGAGAAAGAGGTCATCCCTGATTTGACCGACCACATCCGCCTTTGTGACAACTGTCAGAAATGGTGCCCCA GGTAAGCTCGACGAAACAAAACGAGTTGATCGACATCTAACGCCCACTCAGCGCTGATACAGTCCAATGTGATCG CTGTAAACGCTTCTTCCATATGCGCTGCGTCCAGCCTCCGCTTCTCGCGAAACCTACCCGTGGGTATGGCTGGAC CTGTGCCCCCTGTTCTCGACGGCACGAAGAAGAGGTAGACAGCCATGACGTCCGACCTTCCAGCTCGGCCGCGGG GAAGGCCCATAGGTCGAATGCCCCACCTGCTCGAGGACGAGGTCGGCCCAGGAAGGACAAGACCCTGGCTGAGAA GGAGGAGAATCTTCCTGTGAAACATTTCAAGATGTGGCCTTTCAGATATTTCGGGTGAGGTTTTATCCTCGTTCA CCAAGCTGACTGAATTGACATCACCTTCAATCAGTCAACATACTGTGGCGGAAGATACATTGGGTGAGCGAATTC GCCGCGGGTTTACCAATAACATCACTAACTCCACTATAGACGAAGAAGATCTAATATTCCCTCGTACAGCCACAA GGCACGGTCCAAAATACCAGGCAAATGTCCCTCCAGGACCGGATCCATACAATTACCCCCCAGGTATGCGAATAA GCATAGGGTTTGATACACCATCTCACACCAAATGCTAGACCTCGAAGAGCGTGGTGGTGACAACACAGTGGAAGT TCTCGGCGTTATCAACTCATTGACAGAGTCGGAAGGTACGTATCGAAGTGATCCTTTCAACGGCAATCTATTTAC AACCTTCTTTGACAGTGGCTGAAGGTACGCCGGCCCCATAACTGCAAGCACATGCTTGACTCATATTCCTGGTCT TCCCAGTGGAAGCATGCAAGAAGGGCCTAACCAATGACCCCAAGCTCCGTTACAGCGTTGACTGGTTAACAGAAG TCATCTTCCGTTTCTCTGATGCAGCTCTCGCGGCGCGTCCATTAACGGCTGTCAACATGAAGAATCCCAATCGAA TACAGAAAGTATGCCTTATCCATCCAACATTATATTTTCCCTTCCTAACCTGCGATGCAGTGGAAGAAATACGAA ACGCCGTACACAGATCGTCCCTGGTCACACGACGAAGTCGTCGCCTTTGAAGATGCAGTCGCCCTTCACGGTCCC GAGCTTCGTGCTGCCAAGGACGAAGTGCCAAACCGCTCGATGCCAGAGGTCGTTCGTTTCTTCGGGCACTGGAAG AACTCCAAGCTCAAGGAGGAAAACAAACGTCTCAAAACCTCCGGCCCACCTGCCAAACCAATACATCGCCAATAC CGATCCGCTGCGGAACGAAGCGATGGTCAACGCGTCGGCTTAGGTGACGATGAAGGTTCGATCGTCGCACAACCG TCCAAGTCCCCAAGTTGTGGTGCTTGTCGTACACGTGAATCGCCTCAATGGTGGAAAGCCCCCAAGGGATTGCCA ACGAACATCCTTTGCGAGACATGCGGTACGAACTGGAGGAAGTATGCTGATCTCAATGTCCGTCCAATACGAGAA GAGTCCCTGCCGACTTCGAAGGCGAAGACCTCAGAGAAGCGTGAGGGAACACCCCCGGCTGGTCCTAGCGCCAAG CGGGCAAGAGTACGTTGCTTTTATCAGTGTTCCATGAACTTGAGTGATGACATGTCGATCCAGACTAGCGCTTCT GTGCAGTCAACCCCACCACCAGTGGTGTCTGCAGCCCCGCAGGTCAAATGCTTCGCTTGCCACAAACAGGGTCCG CTCGGCAAGGTCCTTCGATGCAAAAAGTGCCAATTCCGGGCTCATGCAGGTGTGTTGCATTCCCTTTCCATCGCG TAGACTTGGCTCACCCCAATCAGGATGTGTGGGCGCAGTCGTCGGTCCAGCCAAGGTGGACGAGTGGACGTGCGA ACTGTGCGAGAACGAAGAGACCATGGAAGCTTCCATTGTAGGCATCACTTCTCACATCATGCTTTCCTTTCATCC CGGCTCATTTTTCATATCCAGAACTACGACTGCCTCCTTTGCCCTCGTTCTGGACCCGAAGAGAAGAAGCAGAAG CCATGGCCACCAGCCGATACCTTCTTACGAGCATGCAAACCAACAGAAGGACAGGGCTGGACTCATGTCATCTGT GCAGTGTTTTTACCGGAGCTGAGCTTTACGGACGCGTCTCGTTTACGTCTTGTAGAGGGGATGAGTACCATTGCG AAACATCGTTGGACAACGGTAAGTCTTATACTTCCCCCTTGTTTTCCCGTTGACCGAATATTTTCCTGTCAGAAA TGTTGCCTGTGTGGCGAGATGGAGGGAGCGGTAGTGAAATGCAGCGACTGCTATCGAGAATTCCATGTATCTTGC GCCTGGAAAGAGGGGCACAAGTTCGGCTTCGAGATCCAACCAGTGCGTTCTAAAATCCCGGAACGTGGCCCAAAC CACTTACACTTTATTCCAGGTGAAGAGCAGCCGAAGAGATACGACCATCACAACGACGTTCAAGGGTGAATCAGG CTGCATGAGTGCCCTCATCTCTTGTAAGATGCATGATCACAATAAACGTGACATCTATGACATTTGCGAAACCAA CGAGGGTGGCGAGGTGCGCAGTTTGCCTTTTCACCTTTATCAATAATGACAGCATGGCTCATTGAATCTTTACCT CGTTGTTCACAGACCGCGCTGCAGGTGTATTGCCAGGCGTACAAGCAAGCGCAGGTAGGCCAAGCCCATGGCTTG CTGCGCAAAGCCAAACGGCTGGACGCCCTGCTCAACTTACGCGATCCCCACGCACACGGGATGCAACCGGTACAA GAACTCCCCACCACGGAACCCGAATGCAGCAAGTGCAACACGCAATTCTCTCCGGCTTTCTATCCCGTCGTCGAT TTGTCGCCTACAGCTTCCAAAGAGACGTGGCTTTGCCATCGATGTCACTTTGAGGCCAACGTCGAGGTCAACGGC AAGGCAATGGGTATGATGATATCATGAAGCGACGAGGCATGAAGCGACGAGGCACGATGCTCTTTCTTATCAATG AACGCGACACCACTGGCCTCTCCACACGCGTAGTCGTCCAAGGAGTCTGAATGGGCGAATGCAACATGTGAATGC CAAGGAGAAAGGAAGAAGCGAGAGACGAAAGACGAAAGACAACTTGACTATGATGATACAATAAACTACAGCAGG GGATGGGGTGTCGATTGAAATACATCCGGGTGTCTGTCTGTTCTGAAAGAGCGAACGAATCACGAATAACCATAG GAAGGGGGTAGTAGGTAGTAGTAATACGTATGTAATAACGCAAAGCAGGTCCTTACGCGCGAACTCTCTTGTAGA AAAGGACATACGCCTTTTGGTTCTGTAGTTTTCGTTGTTAGTATTATTAGCACGCTGTCTTTTTAATTTATTGAC GACCAATAGGAGTATGACTTACCACGA |
| Length | 4302 |