CopciAB_370755
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_370755 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | GIN1 | Synonyms | 370755 |
| Uniprot id | Functional description | Fungal specific transcription factor domain | |
| Location | scaffold_4:3084179..3088147 | Strand | - |
| Gene length (nt) | 3969 | Transcript length (nt) | 2970 |
| CDS length (nt) | 2970 | Protein length (aa) | 989 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7260660 | 48.4 | 9.718E-276 | 869 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_79697 | 50.1 | 1.19E-268 | 848 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_123182 | 50.4 | 5.56E-268 | 846 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB16844 | 47.1 | 1.862E-245 | 781 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | GIN1 |
|---|---|
| Protein id | CopciAB_370755.T0 |
| Description | Fungal specific transcription factor domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd00067 | GAL4 | IPR001138 | 109 | 147 |
| CDD | cd12148 | fungal_TF_MHR | - | 286 | 691 |
| Pfam | PF00172 | Fungal Zn(2)-Cys(6) binuclear cluster domain | IPR001138 | 113 | 147 |
| Pfam | PF04082 | Fungal specific transcription factor domain | IPR007219 | 296 | 543 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001138 | Zn(2)Cys(6) fungal-type DNA-binding domain |
| IPR007219 | Transcription factor domain, fungi |
| IPR036864 | Zn(2)-C6 fungal-type DNA-binding domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | MF |
| GO:0006355 | regulation of transcription, DNA-templated | BP |
| GO:0008270 | zinc ion binding | MF |
| GO:0003677 | DNA binding | MF |
| GO:0006351 | transcription, DNA-templated | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| B | Fungal specific transcription factor domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| c6 zn |
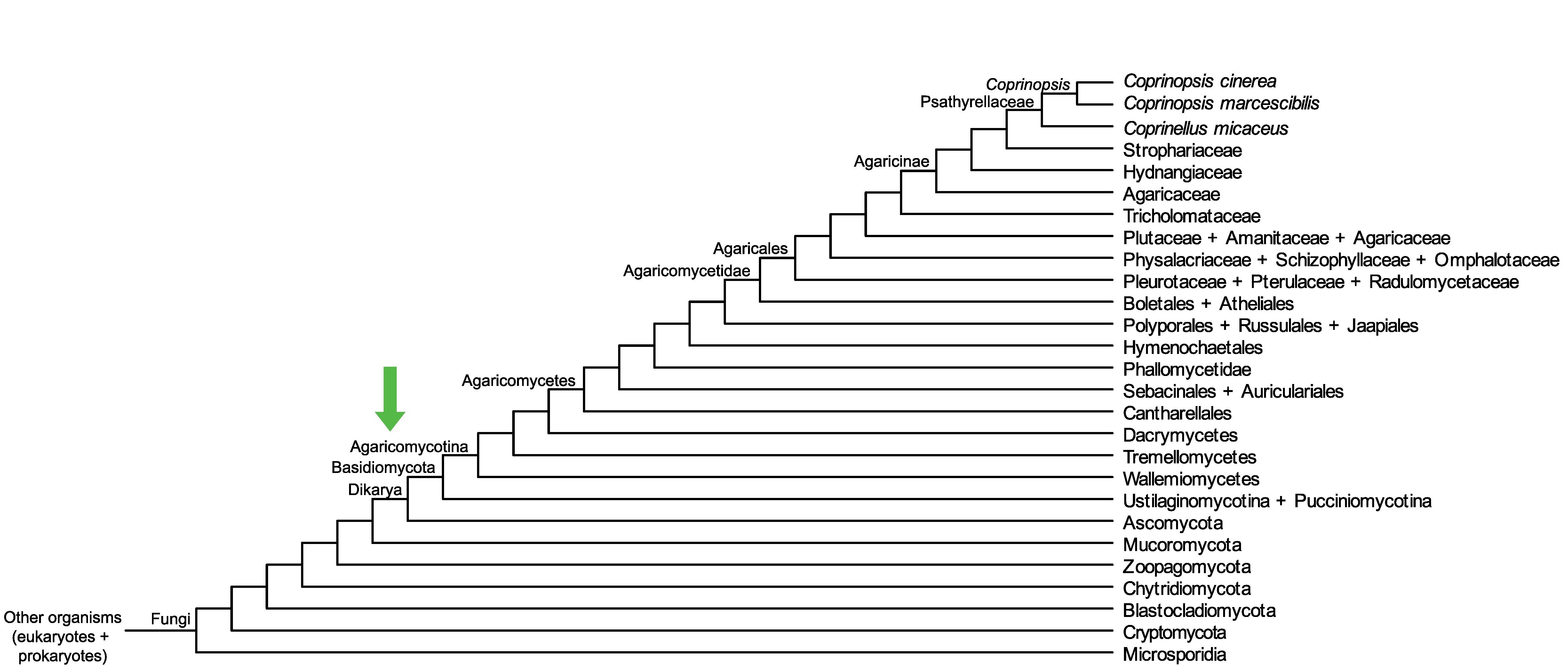
Conservation of CopciAB_370755 across fungi.
Arrow shows the origin of gene family containing CopciAB_370755.
Protein
| Sequence id | CopciAB_370755.T0 |
|---|---|
| Sequence |
>CopciAB_370755.T0 MVGPFAPHCKYSRGFLGVIAKVDTYGEMADKDSCNIANIPLGTDGGHLASWQYRTFTTITPQLSLTPATVHQVMT ADIKMAEPVASTSKLPTGNAEGVLASRTQAKKRRIPGACDICKRKKSKRDSGELPGNRCSNCIQLGLECTHKEVT KTLGSAKGYVESLEMRLEKMDRLLRRLLPGVDLNQELSKPSEPAPSPKPLERNDETEVLGKLNRLTLNPQERRFF GKSSQYDLLQTALDLRQEFTGEGVAYVQSALPMKRPEFWDVPKWMSYNPELDEDQREYAFPDEDLYPSLVNAYFT HINPFLPLLHRPTFEKQIKDGLHLEDSMFARVFLMVCAHGARYSDDPRVFPEGTTSTCSAGWQWFEQVHILRKNL FNRTTLFELQMHALHVLFASSSETPQGVWAQIGMALRLSQEVGGHLRRRRKDGTPPTIEDELWKRAFWVILSLDR LISSSAGRPCGLQDEDFDLDLPLEVDDEYWDQGFVQPEGQPSSVSYFNCYLGLMDILAYAMRLIYPSKRSKYAGK TRSEQQIIAELDTAMNNWMDSLPEHLKWNPGPKNPLFLQQSSALHATYYHIQILIHRPFIPSPRNPIPGAFPSLA ICTNAARSCCHVLEGFTKINFLPTQHFQVIAFTAAVILLLNIWSGKRTGFAPNPRREMEDVSRCMEMLKAAEKRS ASAGRFVDLLTEVASAGDLEAAIKRNASTTKKSKKRSRSDSEDADDASSTSSVTSGSPVLSEARNIAGSRRVSLS QQTSPISPVEPPINFLLPMYGNDLGRLPVYGQFNFADPSNVNAFLGEQLGQIHTSVPTKTSYHFDQPPAAPTSNP PPNPMTLTSDPASAIFTSAADDFAFDNMLKDAMFSCDVIPPSFNPAPQAGTSSSPQTASTSSAGSSSSAASTSAP QLPNLPPLTMSTTDIAALIMQCSGVDYTALLQQLSMLSQLQMQMQLQQPMWDMEMMKLWSSAPANLDRLNDWDAF ISNVGQLTQSQAPT |
| Length | 989 |
Coding
| Sequence id | CopciAB_370755.T0 |
|---|---|
| Sequence |
>CopciAB_370755.T0 ATGGTCGGGCCATTCGCCCCGCACTGCAAGTACTCCCGCGGTTTTTTGGGCGTTATCGCCAAAGTCGATACCTAT GGCGAAATGGCTGACAAAGATAGCTGCAATATTGCGAATATTCCATTGGGTACAGACGGTGGACATCTCGCAAGC TGGCAATATAGGACGTTCACCACGATAACCCCTCAACTATCTCTAACCCCGGCCACAGTACACCAAGTTATGACG GCAGACATCAAGATGGCGGAGCCAGTGGCTTCGACGTCCAAACTTCCAACAGGCAACGCGGAGGGTGTACTAGCG AGTCGTACACAGGCGAAGAAAAGGCGTATTCCAGGCGCATGCGACATCTGTAAACGCAAGAAGAGTAAGCGCGAC AGTGGAGAACTACCCGGAAACAGGTGCAGCAACTGTATTCAATTGGGTCTGGAATGTACACACAAGGAGGTCACA AAGACTCTGGGTTCAGCAAAAGGCTATGTGGAAAGCTTGGAAATGCGGTTAGAAAAGATGGATAGGCTGCTGAGG AGGTTACTGCCGGGAGTTGACTTGAACCAAGAGCTGTCCAAACCTAGCGAGCCCGCCCCTTCGCCAAAACCGTTG GAGAGGAACGACGAGACAGAAGTCCTGGGAAAGCTCAATCGCCTCACTCTCAACCCACAAGAACGACGCTTTTTC GGAAAATCAAGTCAGTATGACTTGCTACAAACGGCTCTTGACCTTCGACAAGAGTTCACTGGAGAGGGTGTTGCG TACGTCCAGTCAGCATTGCCCATGAAGCGGCCAGAGTTCTGGGACGTGCCGAAGTGGATGTCCTACAATCCTGAA CTTGACGAAGACCAACGGGAGTATGCCTTCCCCGACGAAGATCTATACCCTTCCCTTGTCAACGCCTACTTCACC CACATCAATCCATTTCTTCCCCTCTTGCACCGCCCAACTTTTGAAAAACAGATCAAAGATGGACTACATTTGGAG GACTCGATGTTCGCAAGGGTTTTCTTGATGGTTTGTGCCCATGGGGCACGGTATTCCGACGATCCTCGGGTTTTT CCTGAAGGGACTACGTCGACTTGCTCGGCCGGGTGGCAATGGTTCGAACAGGTTCACATTCTTCGGAAGAACCTG TTCAATCGGACGACGCTCTTCGAGTTGCAGATGCACGCACTTCATGTCCTCTTCGCCTCGTCTAGCGAAACGCCA CAAGGTGTTTGGGCCCAAATTGGGATGGCACTGCGATTGTCCCAAGAGGTTGGAGGACACCTACGGAGGAGAAGA AAGGATGGTACGCCTCCGACTATTGAGGACGAGCTATGGAAACGAGCATTCTGGGTGATATTGAGTCTCGATCGT TTAATCAGTTCCAGTGCTGGGCGGCCCTGTGGATTACAAGATGAAGATTTCGATCTGGATCTTCCCCTCGAGGTG GACGACGAATATTGGGATCAGGGTTTCGTTCAGCCAGAGGGACAACCATCGAGCGTATCCTATTTCAATTGCTAT CTTGGCTTGATGGATATCCTGGCATATGCTATGAGATTGATCTATCCCAGCAAACGTTCGAAGTATGCCGGTAAA ACGCGCTCGGAACAGCAGATAATAGCCGAACTCGACACCGCCATGAACAACTGGATGGATTCTTTACCGGAGCAT CTGAAATGGAATCCTGGTCCCAAGAACCCGCTGTTTTTGCAGCAGTCGTCGGCACTTCATGCTACCTATTACCAC ATCCAGATCCTCATTCACAGACCTTTCATTCCCTCGCCGCGGAATCCCATTCCAGGAGCGTTCCCGTCCTTGGCC ATTTGTACGAATGCGGCTCGTTCATGTTGCCATGTCCTCGAAGGGTTCACCAAAATCAACTTCCTTCCTACCCAA CACTTCCAGGTTATCGCCTTTACCGCTGCGGTAATTTTGTTACTGAATATCTGGAGTGGGAAGCGGACTGGGTTT GCACCGAATCCTAGACGTGAGATGGAGGATGTCTCGCGGTGCATGGAAATGCTCAAGGCGGCAGAGAAGAGGTCG GCTTCGGCAGGTCGCTTTGTGGACCTGCTCACTGAAGTCGCATCGGCCGGGGACCTCGAAGCCGCAATAAAGAGA AATGCGAGCACGACGAAGAAGTCCAAGAAGCGTTCACGGTCGGATTCCGAGGACGCCGACGATGCCTCCTCTACT TCGAGTGTCACCTCAGGTTCCCCCGTCCTATCAGAAGCTCGCAATATAGCCGGGTCGCGCCGGGTCTCGTTGAGC CAGCAGACGAGCCCAATATCCCCTGTTGAACCTCCGATCAATTTCCTCCTCCCGATGTATGGGAACGACCTTGGA CGACTACCTGTGTATGGCCAGTTCAATTTTGCAGACCCGTCGAATGTCAACGCCTTCCTGGGCGAGCAGCTGGGT CAGATCCATACCTCCGTTCCAACGAAAACGTCGTATCACTTTGACCAGCCGCCGGCAGCCCCGACGTCAAACCCT CCGCCAAACCCGATGACCCTCACCTCCGACCCTGCTTCCGCCATTTTCACCTCCGCTGCCGACGATTTTGCCTTT GATAATATGCTCAAGGACGCCATGTTCAGCTGTGACGTCATCCCGCCTTCGTTCAACCCAGCTCCTCAGGCTGGC ACTAGCTCTAGTCCTCAAACTGCAAGCACGAGTAGTGCTGGAAGCAGTAGTAGCGCGGCTTCGACGTCTGCTCCA CAGTTGCCGAATTTACCCCCTCTTACCATGTCAACGACTGATATTGCTGCATTGATCATGCAATGTTCGGGAGTG GATTACACTGCGCTTCTTCAGCAGCTGTCCATGTTGAGTCAGTTACAAATGCAAATGCAGCTGCAACAGCCAATG TGGGACATGGAGATGATGAAATTGTGGTCTTCTGCCCCAGCGAACTTGGACAGATTGAATGATTGGGACGCTTTC ATTTCGAATGTCGGGCAACTGACGCAATCTCAAGCTCCTACTTGA |
| Length | 2970 |
Transcript
| Sequence id | CopciAB_370755.T0 |
|---|---|
| Sequence |
>CopciAB_370755.T0 ATGGTCGGGCCATTCGCCCCGCACTGCAAGTACTCCCGCGGTTTTTTGGGCGTTATCGCCAAAGTCGATACCTAT GGCGAAATGGCTGACAAAGATAGCTGCAATATTGCGAATATTCCATTGGGTACAGACGGTGGACATCTCGCAAGC TGGCAATATAGGACGTTCACCACGATAACCCCTCAACTATCTCTAACCCCGGCCACAGTACACCAAGTTATGACG GCAGACATCAAGATGGCGGAGCCAGTGGCTTCGACGTCCAAACTTCCAACAGGCAACGCGGAGGGTGTACTAGCG AGTCGTACACAGGCGAAGAAAAGGCGTATTCCAGGCGCATGCGACATCTGTAAACGCAAGAAGAGTAAGCGCGAC AGTGGAGAACTACCCGGAAACAGGTGCAGCAACTGTATTCAATTGGGTCTGGAATGTACACACAAGGAGGTCACA AAGACTCTGGGTTCAGCAAAAGGCTATGTGGAAAGCTTGGAAATGCGGTTAGAAAAGATGGATAGGCTGCTGAGG AGGTTACTGCCGGGAGTTGACTTGAACCAAGAGCTGTCCAAACCTAGCGAGCCCGCCCCTTCGCCAAAACCGTTG GAGAGGAACGACGAGACAGAAGTCCTGGGAAAGCTCAATCGCCTCACTCTCAACCCACAAGAACGACGCTTTTTC GGAAAATCAAGTCAGTATGACTTGCTACAAACGGCTCTTGACCTTCGACAAGAGTTCACTGGAGAGGGTGTTGCG TACGTCCAGTCAGCATTGCCCATGAAGCGGCCAGAGTTCTGGGACGTGCCGAAGTGGATGTCCTACAATCCTGAA CTTGACGAAGACCAACGGGAGTATGCCTTCCCCGACGAAGATCTATACCCTTCCCTTGTCAACGCCTACTTCACC CACATCAATCCATTTCTTCCCCTCTTGCACCGCCCAACTTTTGAAAAACAGATCAAAGATGGACTACATTTGGAG GACTCGATGTTCGCAAGGGTTTTCTTGATGGTTTGTGCCCATGGGGCACGGTATTCCGACGATCCTCGGGTTTTT CCTGAAGGGACTACGTCGACTTGCTCGGCCGGGTGGCAATGGTTCGAACAGGTTCACATTCTTCGGAAGAACCTG TTCAATCGGACGACGCTCTTCGAGTTGCAGATGCACGCACTTCATGTCCTCTTCGCCTCGTCTAGCGAAACGCCA CAAGGTGTTTGGGCCCAAATTGGGATGGCACTGCGATTGTCCCAAGAGGTTGGAGGACACCTACGGAGGAGAAGA AAGGATGGTACGCCTCCGACTATTGAGGACGAGCTATGGAAACGAGCATTCTGGGTGATATTGAGTCTCGATCGT TTAATCAGTTCCAGTGCTGGGCGGCCCTGTGGATTACAAGATGAAGATTTCGATCTGGATCTTCCCCTCGAGGTG GACGACGAATATTGGGATCAGGGTTTCGTTCAGCCAGAGGGACAACCATCGAGCGTATCCTATTTCAATTGCTAT CTTGGCTTGATGGATATCCTGGCATATGCTATGAGATTGATCTATCCCAGCAAACGTTCGAAGTATGCCGGTAAA ACGCGCTCGGAACAGCAGATAATAGCCGAACTCGACACCGCCATGAACAACTGGATGGATTCTTTACCGGAGCAT CTGAAATGGAATCCTGGTCCCAAGAACCCGCTGTTTTTGCAGCAGTCGTCGGCACTTCATGCTACCTATTACCAC ATCCAGATCCTCATTCACAGACCTTTCATTCCCTCGCCGCGGAATCCCATTCCAGGAGCGTTCCCGTCCTTGGCC ATTTGTACGAATGCGGCTCGTTCATGTTGCCATGTCCTCGAAGGGTTCACCAAAATCAACTTCCTTCCTACCCAA CACTTCCAGGTTATCGCCTTTACCGCTGCGGTAATTTTGTTACTGAATATCTGGAGTGGGAAGCGGACTGGGTTT GCACCGAATCCTAGACGTGAGATGGAGGATGTCTCGCGGTGCATGGAAATGCTCAAGGCGGCAGAGAAGAGGTCG GCTTCGGCAGGTCGCTTTGTGGACCTGCTCACTGAAGTCGCATCGGCCGGGGACCTCGAAGCCGCAATAAAGAGA AATGCGAGCACGACGAAGAAGTCCAAGAAGCGTTCACGGTCGGATTCCGAGGACGCCGACGATGCCTCCTCTACT TCGAGTGTCACCTCAGGTTCCCCCGTCCTATCAGAAGCTCGCAATATAGCCGGGTCGCGCCGGGTCTCGTTGAGC CAGCAGACGAGCCCAATATCCCCTGTTGAACCTCCGATCAATTTCCTCCTCCCGATGTATGGGAACGACCTTGGA CGACTACCTGTGTATGGCCAGTTCAATTTTGCAGACCCGTCGAATGTCAACGCCTTCCTGGGCGAGCAGCTGGGT CAGATCCATACCTCCGTTCCAACGAAAACGTCGTATCACTTTGACCAGCCGCCGGCAGCCCCGACGTCAAACCCT CCGCCAAACCCGATGACCCTCACCTCCGACCCTGCTTCCGCCATTTTCACCTCCGCTGCCGACGATTTTGCCTTT GATAATATGCTCAAGGACGCCATGTTCAGCTGTGACGTCATCCCGCCTTCGTTCAACCCAGCTCCTCAGGCTGGC ACTAGCTCTAGTCCTCAAACTGCAAGCACGAGTAGTGCTGGAAGCAGTAGTAGCGCGGCTTCGACGTCTGCTCCA CAGTTGCCGAATTTACCCCCTCTTACCATGTCAACGACTGATATTGCTGCATTGATCATGCAATGTTCGGGAGTG GATTACACTGCGCTTCTTCAGCAGCTGTCCATGTTGAGTCAGTTACAAATGCAAATGCAGCTGCAACAGCCAATG TGGGACATGGAGATGATGAAATTGTGGTCTTCTGCCCCAGCGAACTTGGACAGATTGAATGATTGGGACGCTTTC ATTTCGAATGTCGGGCAACTGACGCAATCTCAAGCTCCTACTTGA |
| Length | 2970 |
Gene
| Sequence id | CopciAB_370755.T0 |
|---|---|
| Sequence |
>CopciAB_370755.T0 ATGGTCGGGCCATTCGCCCCGCACTGCAAGTACTCCCGCGGTTTTTTGGGCGTTATCGCCAAAGTCGATACCTAT GGCGAAATGGCTGACAAAGATAGCTGCAATATTGCGAATATTCCATTGGGTACAGACGGTGGACGTGGGTTATTT AGGTCTATGAGAGACCCACAACTTGTTCCTTTTGCCCCTAAGATCTCGCAAGCTGGCAATATAGGACGTTCACCA CGATAACCCCTCAACTATCTCTAACCCCGGCCACAGTACACCAAGTTATGACGGCAGACATCAAGATGGCGGAGC CAGTGGCTTCGACGTCCAAACTTCCAACAGGCAACGCGGAGGGTGTACTAGCGAGTCGTACACAGGCGAAGAAAA GGCGTATTCCAGGCGCATGCGACATCTGTAAACGCAAGAAGAGTAAGCGTGGCCCTAATAGTCCCATGACTCGTG CTGATGGTCGCGCTCCTGGCCCTCGTACATCAGTTCGCTGTAGGTTTCCCTATACTTGGGCGGCATCGCCTGTAC ACGAGCCGTGCTCATTCATCACCTTCCTAGGCGACAGTGGAGAACTACCCGGAAACAGGTGCAGCAACTGTATTC AATTGGGTCTGGAATGTACACACAAGGAGGTCACAAAGGTGCGTCCTATCATTATCATCACTGTAATGACGGCAG TGAGTTCTCATGCTGGTCTCTCGACAGACTCTGGGTTCAGCAAAAGGGTACGAGCCCTTTCGGGTTTCATTGCCC TCTGACAGCGGCAATTCCCTCGTTTGGTATGGCTGATTGCCTTTTCTGGATTCGATAGCTATGTGGAAAGCTTGG AAATGCGGTTAGAAAAGATGGATAGGCTGCTGAGGAGGGTGAGTGAGTCGTCGTCTGTGGGTGTGAAATTGATGA TGATGACGGTATACAGTTACTGCCGGGAGTTGACTTGAACCAAGAGCTGTCCAAACCTAGCGAGCCCGCCCCTTC GCCAAAACCGTTGGAGAGGAACGACGAGACAGAAGTCCTGGGAAAGCTCAATCGCCTCACTCTCAACCCACAAGA ACGACGCTTTTTCGGAAAATCAAGGTATTTTCCGGCCTTTACCGCGGCCTTTGCAACGTCTGAACGTCCTCCTCC CATTTAGTCAGTATGACTTGCTACAAACGGCTCTTGACCTTCGACAAGAGTTCACTGGAGAGGGTGTTGCGTACG TCCAGTCAGCATTGCCCATGAAGCGGCCAGAGTTCTGGGACGTGCCGAAGGTAAGTTTTGTCCCTTGGAGAGTAG TTATACGTAACTCACGGAAAACTCAGTGGATGTCCTACAATCCTGAACTTGACGAAGACCAACGGGAGTATGCCT TCCCCGACGAAGATCTATACCCTTCCCTTGTCAACGCCTACTTCACCCACATCAATCCATTTCTTCCCCTCTTGC ACCGCCCAACTTTTGAAAAACAGATCAAAGATGGACTACATTTGGAGGACTCGATGTTCGCAAGGGTTTTCTTGA TGGTTTGTGCCCATGGGGCACGGTATTCCGACGATCCTCGGGTTTTTCCTGAAGGGACTACGTCGACTTGCTCGG CCGGGTGGCAATGGTTCGAACAGGTTCACATTCTTCGGAAGAACCTGTTCAATCGGACGACGCTCTTCGAGTTGC AGATGCACGCAGTTAGTCTTTCACCATCGTGGAACAATCGAGTAGTGAGGCTTATTGACCATTCTCTGCAGCTTC ATGTCCTCTTCGCCTCGTCTAGCGAAACGCCACAAGGTGTTTGGGCCCAAATTGGGATGGCACTGCGATTGTCCC AAGAGGTTGGAGGACACCTACGGAGGAGAAGAAAGGATGGTACGCCTCCGACTATTGAGGACGAGCTATGGAAAC GAGCATTCTGGTACCGTTTTTCGTTCTCTGGTGATGTTTGGCTCAGTGCTTACATTCCGATTACATAGGGTGATA TTGAGTCTCGATCGTTTAATCAGTTCCAGTGCTGGGCGGCCCTGTGGATTACAAGATGAAGAGTACGGTAGTTTC TTCCCGACTTTGAAGCTTCGTTATTTTTACCTTTTCCTCTCAGTTTCGATCTGGATCTTCCCCTCGAGGTGGACG ACGAATATTGGGATCAGGGTTTCGTTCAGCCAGAGGGACAACCATCGAGCGTATCCTATTTCAATTGCTATCTTG GCTTGATGGATATCCTGGCATATGCTATGAGATTGATCGTAAGCAACCTTTCATCCGTAACATTTGGACATTGAA TTGACGTTATTCCGTCGCAACAGTATCCCAGCAAACGTTCGAAGTATGCCGGTAAAACGCGCTCGGAACAGCAGA TAATAGCCGAACTCGACACCGCCATGAACAACTGGATGGATTCTTTACCGGAGCATCGTAAGTGCCGGCCTGATT GGATGAACTTCGCATCTAACTTCGTCCCCAAGTGAAATGGAATCCTGGTCCCAAGAACCCGCTGTTTTTGCAGCA GTCGTCGGCACTTCATGCTACCTATTACCACATCCAGATCCTCATTCACAGACCTTTCATTCCCTCGCCGCGGAA TCCCATTCCAGGAGCGTTCCCGTCCTTGGCCATTTGTACGAATGCGGCTCGTTCATGTTGCCATGTCCTCGAAGG GTTCACCAAAATCAACTTCCTTCCTACCCAACACTTCCAGGTACGTGGAAGAGAGGCACGCCGTTTCGAAATGGA GCTGACGGTTCCCACAAGGTTATCGCCTTTACCGCTGCGGTAATTTTGTTACTGAATATCTGGAGTGGGAAGCGG ACTGGGTTTGCACCGAATCCTAGACGTGAGATGGAGGATGTCTCGCGGTGCATGGAAATGCTCAAGGCGGCAGAG AAGAGGTATGTTCCTGCCTAACGCGTACACTTTTGGGGGGCACTGACCCTTTCCTTGCTTAGGTCGGCTTCGGCA GGTCGCTTTGTGTAAGTGGCTCCTGCTTCCTGGTATTGGTCCCATCTAAATGCCATCCTCACGTTGTAGGGACCT GCTCACTGAAGTCGCATCGGCCGGGGACCTCGAAGCCGCAATAAAGAGAAATGCGAGCACGACGAAGAAGTCCAA GAAGCGTTCACGGTCGGATTCCGAGGACGCCGACGATGCCTCCTCTACTTCGAGTGTCACCTCAGGTTCCCCCGT CCTATCAGAAGCTCGCAATATAGCCGGGTCGCGCCGGGTCTCGTTGAGCCAGCAGACGAGCCCAATATCCCCTGT TGAACCTCCGATCAATTTCCTCCTCCCGATGTATGGGAACGACCTTGGACGACTACCTGTGTATGGCCAGTTCAA TTTTGCAGACCCGTCGAATGTCAACGCCTTCCTGGGCGAGCAGCTGGGTCAGATCCATACCTCCGTTCCAACGAA AACGTCGTATCACTTTGACCAGCCGCCGGCAGCCCCGACGTCAAACCCTCCGCCAAACCCGATGACCCTCACCTC CGACCCTGCTTCCGCCATTTTCACCTCCGCTGCCGACGATTTTGCCTTTGATAATATGCTCAAGGACGCCATGTT CAGCTGTGACGTCATCCCGCCTTCGTTCAACCCAGCTCCTCAGGCTGGCACTAGCTCTAGTCCTCAAACTGCAAG CACGAGTAGTGCTGGAAGCAGTAGTAGCGCGGCTTCGACGTCTGCTCCACAGTTGCCGAATTTACCCCCTCTTAC CATGTCAACGACTGATATTGCTGCATTGATCATGCAATGTTCGGGAGTGGATTACACTGCGCTTCTTCAGCAGCT GTCCATGTTGAGTCAGTTACAAATGCAAATGCAGCTGCAACAGCCAATGTGGGACATGGAGATGATGAAATTGTG GTCTTCTGCCCCAGCGAACTTGGAGTAAGTCTTGACTTGTGGCTGTCAGCTGCGAAGAGCTAATCCAATCGCAGC AGATTGAATGATTGGGACGCTTTCATTTCGAATGTCGGGCAACTGACGCAATCTCAAGCTCCTACTTGA |
| Length | 3969 |