CopciAB_374032
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_374032 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 374032 |
| Uniprot id | Functional description | Protein kinase domain | |
| Location | scaffold_13:297277..298939 | Strand | - |
| Gene length (nt) | 1663 | Transcript length (nt) | 1600 |
| CDS length (nt) | 1476 | Protein length (aa) | 491 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN10325_srpkD |
| Neurospora crassa | stk-35 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29753 | 47.7 | 1.57E-129 | 420 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_152487 | 45.6 | 1.549E-123 | 402 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_102163 | 44.9 | 1.055E-118 | 388 |
| Grifola frondosa | Grifr_OBZ66636 | 47.2 | 1.415E-105 | 350 |
| Lentinula edodes NBRC 111202 | Lenedo1_1125812 | 42.6 | 1.651E-104 | 347 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_19085 | 42.6 | 1.638E-104 | 347 |
| Lentinula edodes B17 | Lened_B_1_1_8355 | 42.4 | 3.075E-103 | 343 |
| Agrocybe aegerita | Agrae_CAA7268662 | 45.8 | 1.81E-76 | 265 |
| Auricularia subglabra | Aurde3_1_1179570 | 32.7 | 2.58E-52 | 194 |
| Schizophyllum commune H4-8 | Schco3_2716838 | 27.1 | 8.089E-30 | 125 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_374032.T0 |
| Description | Protein kinase domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF06293 | Lipopolysaccharide kinase (Kdo/WaaP) family | - | 199 | 243 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 299 | 487 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008266 | Tyrosine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein kinase domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
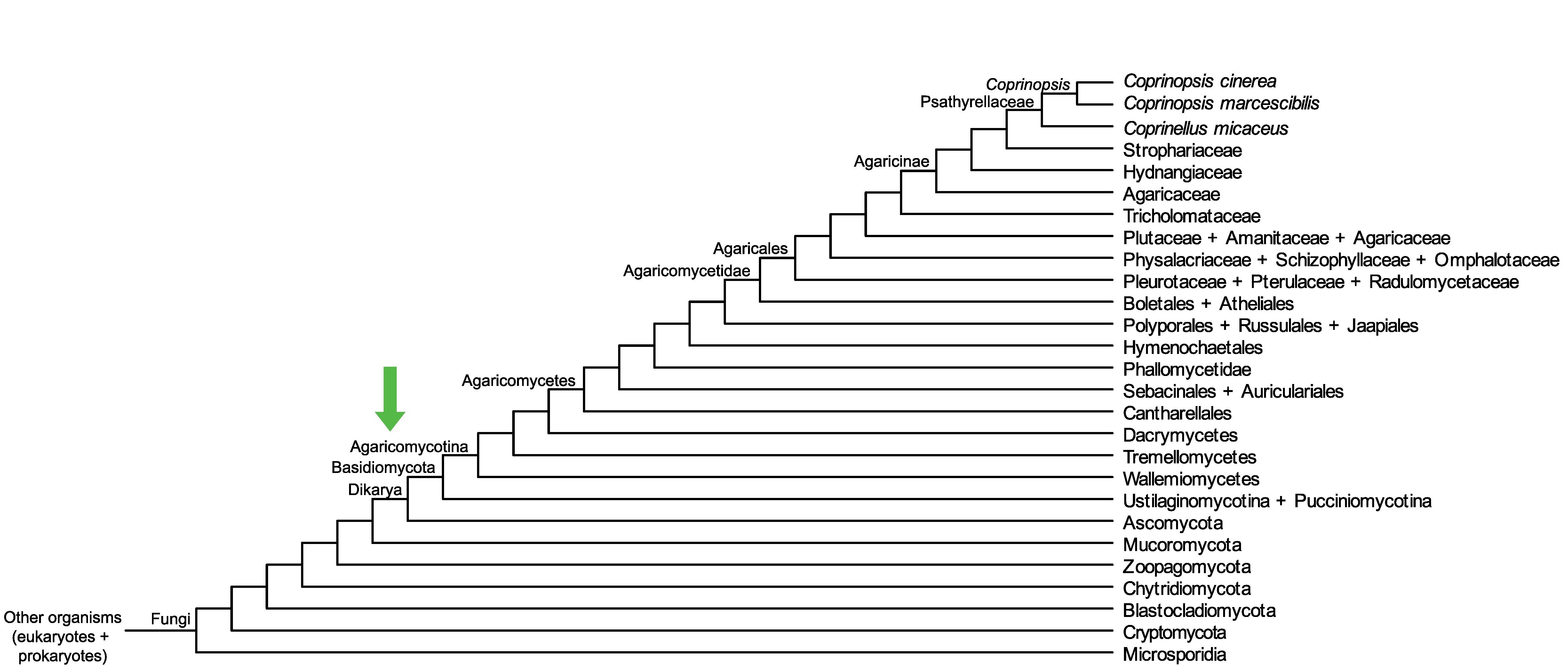
Conservation of CopciAB_374032 across fungi.
Arrow shows the origin of gene family containing CopciAB_374032.
Protein
| Sequence id | CopciAB_374032.T0 |
|---|---|
| Sequence |
>CopciAB_374032.T0 MHPSCPLVARIQLSSYLFRIKRRPFPATKRFSSFMASPYPPISIGGPGGDYRYRPSNLDDVENVENYVEGVYHPV SIGDTLANGRYKLIHKLGFGGSSTVWLARDQQPDSTTPDAASLVALKVMAAHFSDRPPSAMQELLVPEKLHALFS ESRHPVKDHIHRIHRHFTETGPNGTHLCLVAPFCGPSLLAVADNLSYRYHRRIEGRVARRLAKQAALVIERMHSA GMVHGDITTSNMLFRLKESVQAMSDDAICAQLGEPVTEEVITASGDPPGPDAPRELVESIQASRFIEFSLVHDSI LVSDFGQSFLVDAMPEDYHPATTIHCTPPEVRFDQTISFASDVWMLACTLFEIRAGFSLFDSFLRSENLVWRQAV EMLGKLPDPWWNQWDARKVWFAESDGEPKPEEVQKEEGVLLVARKCTIRDKLELIGKKEEPPRGKDWPMTEPDET VMEEEEIKLLQDLLQKMLRYRPEERISMAEVIQHPWFNYKD |
| Length | 491 |
Coding
| Sequence id | CopciAB_374032.T0 |
|---|---|
| Sequence |
>CopciAB_374032.T0 ATGCATCCATCATGTCCGCTCGTTGCCCGAATCCAGCTCTCTTCATATCTTTTTAGGATCAAACGGAGACCTTTT CCCGCAACAAAACGGTTCTCATCCTTCATGGCTAGCCCTTACCCGCCAATTTCTATAGGAGGCCCTGGTGGCGAC TACCGCTACCGTCCTTCGAACCTCGACGACGTCGAAAATGTTGAAAACTACGTCGAAGGAGTCTACCATCCCGTC TCTATTGGCGACACCCTCGCCAACGGCCGCTATAAACTCATTCACAAGCTGGGCTTTGGCGGTTCCTCCACCGTA TGGCTTGCTCGGGACCAACAACCAGATTCTACCACTCCAGACGCGGCCAGCTTGGTGGCGCTGAAAGTCATGGCC GCGCATTTTTCAGACAGGCCCCCTTCGGCAATGCAGGAGCTTCTCGTTCCTGAGAAACTGCACGCCTTGTTTTCC GAGTCTCGTCATCCCGTAAAAGACCACATCCACCGTATCCACAGGCATTTCACCGAAACTGGACCCAACGGGACG CATCTCTGCTTGGTGGCGCCCTTTTGTGGCCCGAGTCTCCTTGCTGTTGCCGATAACCTCTCGTATCGGTACCAC AGGAGAATCGAAGGGCGCGTTGCACGACGGTTGGCGAAACAAGCTGCACTCGTCATCGAGCGCATGCATAGTGCC GGCATGGTCCATGGAGATATCACCACGTCAAATATGCTGTTCAGGCTCAAAGAAAGCGTGCAGGCCATGTCAGAC GACGCGATCTGCGCACAGCTCGGAGAACCCGTTACGGAGGAGGTGATCACTGCAAGTGGCGATCCTCCAGGGCCT GACGCGCCCCGTGAACTGGTGGAATCAATCCAAGCTTCCCGCTTTATAGAATTCTCCTTGGTGCATGACAGCATC CTTGTCTCAGACTTTGGGCAGTCCTTCCTCGTTGACGCCATGCCCGAAGATTACCATCCAGCCACGACCATCCAC TGCACCCCTCCGGAAGTCCGCTTCGATCAAACCATTTCCTTCGCCTCCGACGTTTGGATGCTCGCTTGCACCTTG TTCGAGATCCGCGCTGGATTCTCTCTCTTCGACTCATTCCTCAGAAGCGAGAATCTCGTGTGGAGGCAGGCGGTC GAGATGCTAGGCAAGCTACCAGATCCCTGGTGGAACCAGTGGGACGCTCGCAAAGTTTGGTTTGCCGAGTCCGAC GGAGAACCCAAACCGGAGGAGGTCCAGAAGGAGGAGGGCGTTCTGCTGGTTGCGCGCAAGTGCACCATTCGTGAC AAGCTGGAGCTCATCGGCAAGAAGGAGGAGCCTCCTCGAGGAAAGGATTGGCCGATGACGGAACCGGATGAGACG GTCATGGAGGAGGAGGAAATCAAGCTACTCCAGGATCTGCTGCAAAAGATGCTCAGGTACCGTCCGGAGGAGCGG ATTAGCATGGCGGAGGTGATTCAACATCCTTGGTTTAATTATAAAGACTAG |
| Length | 1476 |
Transcript
| Sequence id | CopciAB_374032.T0 |
|---|---|
| Sequence |
>CopciAB_374032.T0 AGCGCTTAACGCCGAATGCATCCATCATGTCCGCTCGTTGCCCGAATCCAGCTCTCTTCATATCTTTTTAGGATC AAACGGAGACCTTTTCCCGCAACAAAACGGTTCTCATCCTTCATGGCTAGCCCTTACCCGCCAATTTCTATAGGA GGCCCTGGTGGCGACTACCGCTACCGTCCTTCGAACCTCGACGACGTCGAAAATGTTGAAAACTACGTCGAAGGA GTCTACCATCCCGTCTCTATTGGCGACACCCTCGCCAACGGCCGCTATAAACTCATTCACAAGCTGGGCTTTGGC GGTTCCTCCACCGTATGGCTTGCTCGGGACCAACAACCAGATTCTACCACTCCAGACGCGGCCAGCTTGGTGGCG CTGAAAGTCATGGCCGCGCATTTTTCAGACAGGCCCCCTTCGGCAATGCAGGAGCTTCTCGTTCCTGAGAAACTG CACGCCTTGTTTTCCGAGTCTCGTCATCCCGTAAAAGACCACATCCACCGTATCCACAGGCATTTCACCGAAACT GGACCCAACGGGACGCATCTCTGCTTGGTGGCGCCCTTTTGTGGCCCGAGTCTCCTTGCTGTTGCCGATAACCTC TCGTATCGGTACCACAGGAGAATCGAAGGGCGCGTTGCACGACGGTTGGCGAAACAAGCTGCACTCGTCATCGAG CGCATGCATAGTGCCGGCATGGTCCATGGAGATATCACCACGTCAAATATGCTGTTCAGGCTCAAAGAAAGCGTG CAGGCCATGTCAGACGACGCGATCTGCGCACAGCTCGGAGAACCCGTTACGGAGGAGGTGATCACTGCAAGTGGC GATCCTCCAGGGCCTGACGCGCCCCGTGAACTGGTGGAATCAATCCAAGCTTCCCGCTTTATAGAATTCTCCTTG GTGCATGACAGCATCCTTGTCTCAGACTTTGGGCAGTCCTTCCTCGTTGACGCCATGCCCGAAGATTACCATCCA GCCACGACCATCCACTGCACCCCTCCGGAAGTCCGCTTCGATCAAACCATTTCCTTCGCCTCCGACGTTTGGATG CTCGCTTGCACCTTGTTCGAGATCCGCGCTGGATTCTCTCTCTTCGACTCATTCCTCAGAAGCGAGAATCTCGTG TGGAGGCAGGCGGTCGAGATGCTAGGCAAGCTACCAGATCCCTGGTGGAACCAGTGGGACGCTCGCAAAGTTTGG TTTGCCGAGTCCGACGGAGAACCCAAACCGGAGGAGGTCCAGAAGGAGGAGGGCGTTCTGCTGGTTGCGCGCAAG TGCACCATTCGTGACAAGCTGGAGCTCATCGGCAAGAAGGAGGAGCCTCCTCGAGGAAAGGATTGGCCGATGACG GAACCGGATGAGACGGTCATGGAGGAGGAGGAAATCAAGCTACTCCAGGATCTGCTGCAAAAGATGCTCAGGTAC CGTCCGGAGGAGCGGATTAGCATGGCGGAGGTGATTCAACATCCTTGGTTTAATTATAAAGACTAGGTTTATCGA TACACCGAACTCAACTGTGGAAGACTAGTCTCGCCTACCATTGTTCTCCTCCCAAGTGTACGCCTCCTCTTCCAC GCCTATTGAAGCTGTGTCATTCGTC |
| Length | 1600 |
Gene
| Sequence id | CopciAB_374032.T0 |
|---|---|
| Sequence |
>CopciAB_374032.T0 AGCGCTTAACGCCGAATGCATCCATCATGTCCGCTCGTTGCCCGAATCCAGCTCTCTTCATATCTTTTTAGGATC AAACGGAGACCTTTTCCCGCAACAAAACGGTTCTCATCCTTCATGGCTAGCCCTTACCCGCCAATTTCTATAGGA GGCCCTGGTGGCGACTACCGCTACCGTCCTTCGAACCTCGACGACGTCGAAAATGTTGAAAACTACGTCGAAGGA GTCTACCATCCCGTCTCTATTGGCGACACCCTCGCCAACGGCCGCTATAAACTCATTCACAAGCTGGGCTTTGGC GGTTCCTCCACCGTATGGCTTGCTCGGGACCAACAACCAGATTCTACCACTCCAGACGCGGCCAGCTTGGTGGCG CTGAAAGTCATGGCCGCGCATTTTTCAGACAGGCCCCCTTCGGCAATGCAGGAGCTTCTCGTTCCTGAGAAACTG CACGCCTTGTTTTCCGAGTCTCGTCATCCCGTAAAAGACCACATCCACCGTATCCACAGGCATTTCACCGAAACT GGACCCAACGGGACGCATCTCTGCTTGGTGGCGCCCTTTTGTGGCCCGAGTCTCCTTGCTGTTGCCGATAACCTC TCGTATCGGTACCACAGGAGAATCGAAGGGCGCGTTGCACGACGGTTGGCGAAACAAGCTGCACTCGTCATCGAG CGCATGCATAGTGCCGGCATGGTCCATGGAGGTTCGTGCTTTGACGTCTCTGTGCGACCTATCCTCACCCAGTGT CTGTTTCGCAACTTTACAGATATCACCACGTCAAATATGCTGTTCAGGCTCAAAGAAAGCGTGCAGGCCATGTCA GACGACGCGATCTGCGCACAGCTCGGAGAACCCGTTACGGAGGAGGTGATCACTGCAAGTGGCGATCCTCCAGGG CCTGACGCGCCCCGTGAACTGGTGGAATCAATCCAAGCTTCCCGCTTTATAGAATTCTCCTTGGTGCATGACAGC ATCCTTGTCTCAGACTTTGGGCAGTCCTTCCTCGTTGACGCCATGCCCGAAGATTACCATCCAGCCACGACCATC CACTGCACCCCTCCGGAAGTCCGCTTCGATCAAACCATTTCCTTCGCCTCCGACGTTTGGATGCTCGCTTGCACC TTGTTCGAGATCCGCGCTGGATTCTCTCTCTTCGACTCATTCCTCAGAAGCGAGAATCTCGTGTGGAGGCAGGCG GTCGAGATGCTAGGCAAGCTACCAGATCCCTGGTGGAACCAGTGGGACGCTCGCAAAGTTTGGTTTGCCGAGTCC GACGGAGAACCCAAACCGGAGGAGGTCCAGAAGGAGGAGGGCGTTCTGCTGGTTGCGCGCAAGTGCACCATTCGT GACAAGCTGGAGCTCATCGGCAAGAAGGAGGAGCCTCCTCGAGGAAAGGATTGGCCGATGACGGAACCGGATGAG ACGGTCATGGAGGAGGAGGAAATCAAGCTACTCCAGGATCTGCTGCAAAAGATGCTCAGGTACCGTCCGGAGGAG CGGATTAGCATGGCGGAGGTGATTCAACATCCTTGGTTTAATTATAAAGACTAGGTTTATCGATACACCGAACTC AACTGTGGAAGACTAGTCTCGCCTACCATTGTTCTCCTCCCAAGTGTACGCCTCCTCTTCCACGCCTATTGAAGC TGTGTCATTCGTC |
| Length | 1663 |