CopciAB_374043
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_374043 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | BUD32 | Synonyms | 374043 |
| Uniprot id | Functional description | Lipopolysaccharide kinase (Kdo/WaaP) family | |
| Location | scaffold_13:381466..383939 | Strand | + |
| Gene length (nt) | 2474 | Transcript length (nt) | 1207 |
| CDS length (nt) | 927 | Protein length (aa) | 308 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN2513_pipA |
| Schizosaccharomyces pombe | SPAP27G11.07c |
| Saccharomyces cerevisiae | YGR262C_BUD32 |
| Fusarium graminearum | BUD32 |
| Candida albicans | BUD32 |
| Neurospora crassa | prk-11 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21769 | 56.8 | 4.506E-94 | 308 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_165856 | 55.5 | 9.228E-92 | 301 |
| Schizophyllum commune H4-8 | Schco3_2629818 | 54 | 2.991E-89 | 294 |
| Agrocybe aegerita | Agrae_CAA7264457 | 53.3 | 6.102E-89 | 293 |
| Lentinula edodes NBRC 111202 | Lenedo1_1164106 | 54.1 | 2.866E-88 | 291 |
| Lentinula edodes B17 | Lened_B_1_1_7469 | 61.1 | 2.911E-85 | 282 |
| Auricularia subglabra | Aurde3_1_1357518 | 48.4 | 2.739E-75 | 254 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1079844 | 57.2 | 1.985E-71 | 242 |
| Flammulina velutipes | Flave_chr09AA00296 | 58.5 | 7.464E-65 | 223 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_1669 | 58 | 9.298E-63 | 217 |
| Pleurotus ostreatus PC9 | PleosPC9_1_90152 | 56.3 | 7.608E-52 | 185 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | BUD32 |
|---|---|
| Protein id | CopciAB_374043.T0 |
| Description | Lipopolysaccharide kinase (Kdo/WaaP) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF06293 | Lipopolysaccharide kinase (Kdo/WaaP) family | - | 186 | 274 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR022495 | Serine/threonine-protein kinase Bud32 |
| IPR008266 | Tyrosine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K08851 |
EggNOG
| COG category | Description |
|---|---|
| T | Lipopolysaccharide kinase (Kdo/WaaP) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
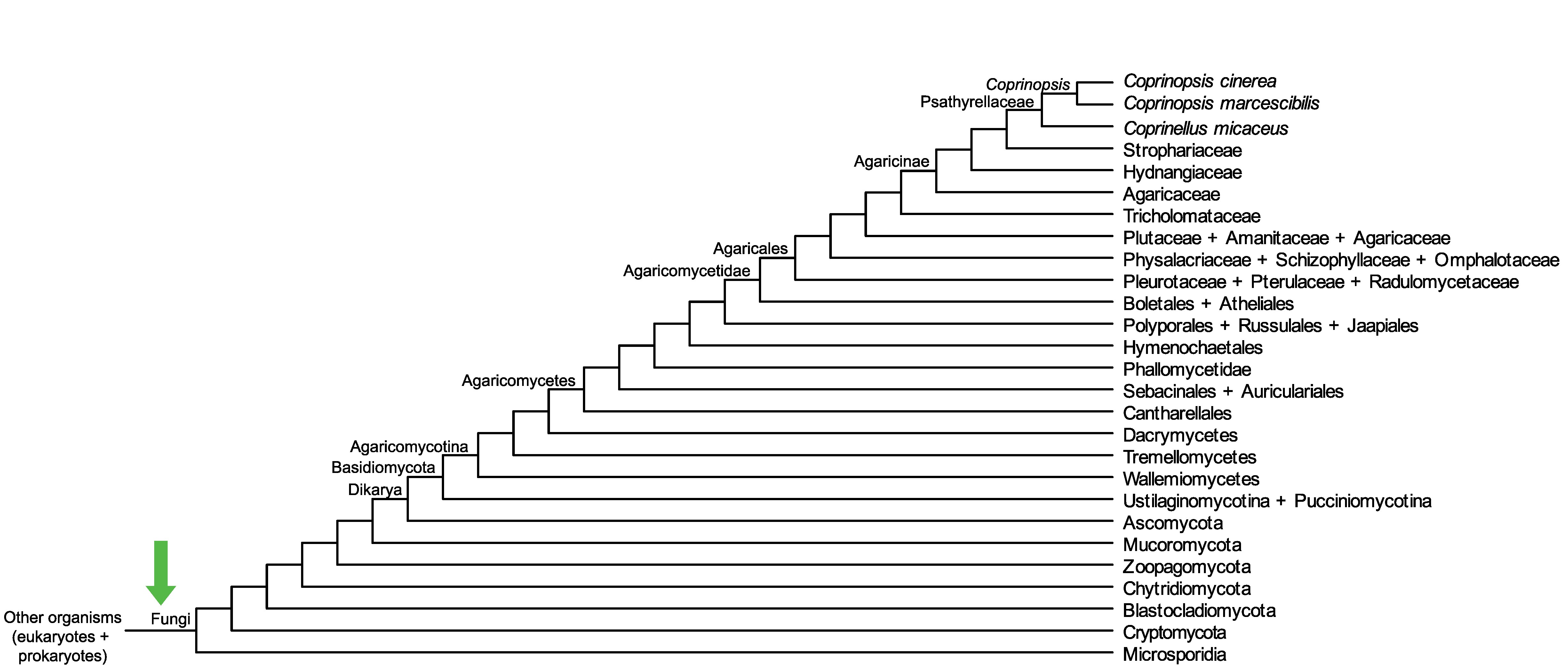
Conservation of CopciAB_374043 across fungi.
Arrow shows the origin of gene family containing CopciAB_374043.
Protein
| Sequence id | CopciAB_374043.T0 |
|---|---|
| Sequence |
>CopciAB_374043.T0 MLSSHSQRPLPASQQLIANAQAISQGAEARIYKTSLTSSNAFSPSTPSAPTTTTAATTTTTTESSQPPAPQSPPI LLKHRFKKKYRHDVLDVSLTKQRVAGEARALLKCLRSGVNVPQILMVDAAEGILGIEWIDGHSVRQLLPGGEEEE EEGEEQREEGQQDAPDSIDEAEERLKKEYGVDIDTLLTLIGTELSKLHKADIIHGDLTTSNMMVRRKAGGGGGGG GGGGAEVLLIDFGLSYTSTLTEDKAVDLYVLERAFASTHPDSERLFGAVLGAYERGLGPKEWHTVKKRLDEVRLR GRKRSMVG |
| Length | 308 |
Coding
| Sequence id | CopciAB_374043.T0 |
|---|---|
| Sequence |
>CopciAB_374043.T0 ATGCTCTCCTCGCACTCCCAACGGCCCCTCCCGGCATCGCAACAACTCATCGCAAACGCGCAAGCTATCTCGCAG GGTGCTGAAGCTAGAATCTACAAAACGTCTTTAACTTCTTCGAACGCCTTCTCCCCGTCCACACCCTCGGCTCCA ACGACGACAACAGCGGCCACCACCACAACGACAACAGAATCATCACAACCCCCAGCACCACAATCCCCTCCCATC CTCCTCAAACACCGCTTCAAAAAAAAGTACCGACACGACGTGTTGGATGTGTCGTTGACGAAGCAGAGGGTGGCC GGCGAGGCTCGGGCGTTGTTGAAGTGTTTGCGATCCGGCGTCAACGTCCCGCAAATCCTCATGGTGGACGCAGCG GAGGGTATCCTCGGTATCGAATGGATAGATGGCCACAGTGTGCGGCAATTACTACCCGGTGGCGAGGAGGAAGAA GAGGAAGGCGAAGAACAAAGAGAAGAAGGACAGCAAGACGCACCAGACTCGATAGATGAAGCTGAGGAAAGGTTG AAGAAGGAGTATGGTGTTGATATCGACACCCTCCTAACCCTCATCGGCACCGAACTTTCCAAACTCCACAAAGCG GATATCATACATGGAGACTTGACCACGTCGAATATGATGGTGCGGCGGAAGGCTGGTGGAGGAGGAGGAGGAGGA GGAGGAGGAGGAGCTGAAGTGCTCCTAATAGACTTTGGCCTCTCCTACACCTCTACTCTAACCGAAGACAAAGCG GTCGACTTGTACGTCCTCGAGCGCGCGTTTGCGTCTACGCACCCTGATTCGGAGCGGTTGTTTGGTGCTGTGTTG GGGGCTTATGAGCGGGGGTTGGGCCCGAAGGAATGGCACACTGTTAAGAAGAGGTTGGATGAAGTGAGGTTGAGA GGGCGGAAGAGGAGTATGGTTGGGTAG |
| Length | 927 |
Transcript
| Sequence id | CopciAB_374043.T0 |
|---|---|
| Sequence |
>CopciAB_374043.T0 GTTACATCCTTTCCCAGTCCATAAGCTCTACTCGTCGCTCTACTCTTAAGGAGAAAGGCTTTTCTTCGTGGGCTA TAAAAGCAAATGCTCTCCTCGCACTCCCAACGGCCCCTCCCGGCATCGCAACAACTCATCGCAAACGCGCAAGCT ATCTCGCAGGGTGCTGAAGCTAGAATCTACAAAACGTCTTTAACTTCTTCGAACGCCTTCTCCCCGTCCACACCC TCGGCTCCAACGACGACAACAGCGGCCACCACCACAACGACAACAGAATCATCACAACCCCCAGCACCACAATCC CCTCCCATCCTCCTCAAACACCGCTTCAAAAAAAAGTACCGACACGACGTGTTGGATGTGTCGTTGACGAAGCAG AGGGTGGCCGGCGAGGCTCGGGCGTTGTTGAAGTGTTTGCGATCCGGCGTCAACGTCCCGCAAATCCTCATGGTG GACGCAGCGGAGGGTATCCTCGGTATCGAATGGATAGATGGCCACAGTGTGCGGCAATTACTACCCGGTGGCGAG GAGGAAGAAGAGGAAGGCGAAGAACAAAGAGAAGAAGGACAGCAAGACGCACCAGACTCGATAGATGAAGCTGAG GAAAGGTTGAAGAAGGAGTATGGTGTTGATATCGACACCCTCCTAACCCTCATCGGCACCGAACTTTCCAAACTC CACAAAGCGGATATCATACATGGAGACTTGACCACGTCGAATATGATGGTGCGGCGGAAGGCTGGTGGAGGAGGA GGAGGAGGAGGAGGAGGAGGAGCTGAAGTGCTCCTAATAGACTTTGGCCTCTCCTACACCTCTACTCTAACCGAA GACAAAGCGGTCGACTTGTACGTCCTCGAGCGCGCGTTTGCGTCTACGCACCCTGATTCGGAGCGGTTGTTTGGT GCTGTGTTGGGGGCTTATGAGCGGGGGTTGGGCCCGAAGGAATGGCACACTGTTAAGAAGAGGTTGGATGAAGTG AGGTTGAGAGGGCGGAAGAGGAGTATGGTTGGGTAGGCGAGGTGTTTCTGTGTTATGGTTGGGTAGACGAGGTGT TTCTGTGTATGGGGTTGGGTAGACGAGCTGGTTTCTGTTTTATCTATTTTTTTTTCGGAGCGAAACGTTGTAGTA TGGACGAGTCGGATGTGTATGTAGTGCAAAACGGTTACAGCGTATGGGTGTAGCGCAATGTACAAAAACGTTGTA TGTGTGT |
| Length | 1207 |
Gene
| Sequence id | CopciAB_374043.T0 |
|---|---|
| Sequence |
>CopciAB_374043.T0 GTTACATCCTTTCCCAGTCCATAAGCTCTACTCGTCGCTCTACTCTTAAGGAGAAAGGCTTTTCTTCGTGGGCTA TAAAAGCAAATGCTCTCCTCGCACTCCCAACGGCCCCTCCCGGCATCGCAACAACTCATCGCAAACGCGCAAGCT ATCTCGCAGGGTGCTGAAGCTGCAAGTCCTTCCTCCTTGTCGTTTAAGGATGCGTTGAATAGCTTATACTCAGGG ATCCCGTTATACCACTTTACTTGCCTTGGTGTCGTCACCCTGGTTCTGTCTATCGACAAACGATAAACGTCCGGG GCGAATCCTCCTATCCCATGACCGTCGATCATAACAGAGAATCTACAAAACGTCTTTAACTTCTTCGAACGCCTT CTCCCCGTCCACACCCTCGGCTCCAACGACGACAACAGCGGCCACCACCACAACGACAACAGAATCATCACAACC CCCAGCACCACAATCCCCTCCCATCCTCCTCAAACACCGCTTCAAAAAAAAGTACCGACACGACGTGTTGGATGT GTCGTTGACGAAGCAGAGGGTGGCCGGCGAGGCTCGGGCGTTGTTGAAGTGTTTGCGGTTAGTGCTCAATCGCGG TTTCTCTTTCTCTTTCTTTCGTTCTTTGGCCATTCTTCGTTGGCGGTCCCGCTTCGATGATCTCCTCTGTTCGAC GACGACTCCCCATCTGCTCAACGCCTACTACCTCAGTCGCTATCCGTTATGAGGGAACCAGTGACGACTCGCTAA CCCCCACCCCCATCGTCATCTAGATCCGGCGTCAACGTCCCGCAAATCCTCATGGTGGACGCAGCGGAGGGTATC CTCGGTATCGAATGGATAGATGGCCACAGTGTGCGGCAATTACTACCCGGTGGCGAGGAGGAAGAAGAGGAAGGC GAAGAACAAAGAGAAGGTGAGGAATAGTTTACTGTTACACGCCTTTCGAGGTGGATGGAACTAACGCCGGGGGTT GAAATCGAAGAAGGACAGCAAGACGCACCAGACTCGATAGATGAAGCTGAGGAAAGGTTGAAGAAGGAGTATGGT GTTGATATCGGTGTGTGTTACTTCTTTCCTTCCATCACGTCCTCTCGAACTTACCACGAAATCCATCTCTACCAC GCCCACCAAATCACGACCCACAGACACCCTCCTAACCCTCATCGGCACCGAACTTTCCAAACTCCACAAAGCGGA TATCATACATGGAGACTTGACCACGTCGAATATGATGGTGCGGCGGAAGGCTGGTGGAGGAGGAGGAGGAGGAGG AGGAGGAGGAGCTGAAGTGGTACGTAGTTTCTCTGCATGGTCGTTTGTGCCAGTTATTCTCTTCTTGGTCATTAT CTTCTCTTCTCAGTTTGCCCCCGTCCCCGTCCTTGTCCACTCTTTCGTTATCCTTCCATCTTCTGCCCTCGCCCT CTGCCCTCCTCTCCTCTCCCGTCCCTATCTCTACATCTCTACATACAATCCGGCTCCCCCTAACACCCTCCCTCT CCTCCCCAACAGCTCCTAATAGACTTTGGCCTCTCCTACACCTCTACTCTAACCGAAGACAAAGCGGTCGACTTG TACGTCCTCGAGCGCGCGTTTGCGTCTACGCACCCTGATTCGGAGCGGTTGTTTGGTGCTGTGTTGGGGGCTTAT GAGCGGGGGTTGGGTGAGTGTTTTTTTATGTTTTTTTTTTTTTTTGTGGATTCTATTTTTTCTGGATTTAATTGA TTTTGGGGCTCGGGGGGTTTGAGGTGCGAGTTTTGGAAATGATTGGGTTTGAAAAAAAATCATGGGGGTTTGACG AGCGTAAATCACGGGGTTTGACGAGCGTGGCGTTGTCCTCGTTGCGAATTCTTGGGCTCTTATGTTCGCGTCCGG GTCTAGGATCCGGATTTATTTTGGTGACGCGTTGTGATCGTTTGTCCGTTTCTCTTGGAGCGAACTGGGGTGGGT GGGTAGGTGGGGTGGGTGGGTTGGAGGATGGGTTGACGTGCTTCGTGCTCCTGGACCTTGACTCGATGAACGACG ATCGCCTGAACCGCGAACGACCTAACGGACTAGCGCTTTCTGAATTTAAAGTCTCGCTAACCCTCTACATTCAAA CTCCCTCGAACCCCTCTACATTCGAACTCTCTCTAAAACCCGCAGTCGTAAACGGACTGACTAACAAACCCTCCT TTTTCTTTGGATGTACATTGTAGGCCCGAAGGAATGGCACACTGTTAAGAAGAGGTTGGATGAAGTGAGGTTGAG AGGGCGGAAGAGGAGTATGGTTGGGTAGGCGAGGTGTTTCTGTGTTATGGTTGGGTAGACGAGGTGTTTCTGTGT ATGGGGTTGGGTAGACGAGCTGGTTTCTGTTTTATCTATTTTTTTTTCGGAGCGAAACGTTGTAGTATGGACGAG TCGGATGTGTATGTAGTGCAAAACGGTTACAGCGTATGGGTGTAGCGCAATGTACAAAAACGTTGTATGTGTGT |
| Length | 2474 |