CopciAB_374320
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_374320 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 374320 |
| Uniprot id | Functional description | protein Coprinopsis cinerea okayama7 130 | |
| Location | scaffold_9:1200743..1202097 | Strand | + |
| Gene length (nt) | 1355 | Transcript length (nt) | 1235 |
| CDS length (nt) | 948 | Protein length (aa) | 315 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Grifola frondosa | Grifr_OBZ77296 | 38.1 | 4.683E-64 | 221 |
| Lentinula edodes NBRC 111202 | Lenedo1_1169066 | 33 | 8.274E-44 | 162 |
| Agrocybe aegerita | Agrae_CAA7271258 | 39.1 | 2.483E-37 | 143 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_6814 | 33.3 | 2.831E-31 | 125 |
| Lentinula edodes B17 | Lened_B_1_1_3721 | 33.8 | 5.106E-24 | 103 |
| Auricularia subglabra | Aurde3_1_1229111 | 30.3 | 1.012E-21 | 97 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_109484 | 29.6 | 2.232E-17 | 83 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191477 | 31.1 | 0 | 44 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114440 | 31.1 | 0 | 44 |
| Pleurotus ostreatus PC9 | PleosPC9_1_121819 | 45.2 | 0.0001 | 43 |
| Pleurotus eryngii ATCC 90797 | Pleery1_592776 | 45.2 | 0.0003 | 42 |
| Pleurotus ostreatus PC15 | PleosPC15_2_164797 | 44.4 | 0.0002 | 42 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_374320.T0 |
| Description | protein Coprinopsis cinerea okayama7 130 |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| No records | |||||
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| No records | |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | protein Coprinopsis cinerea okayama7 130 |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
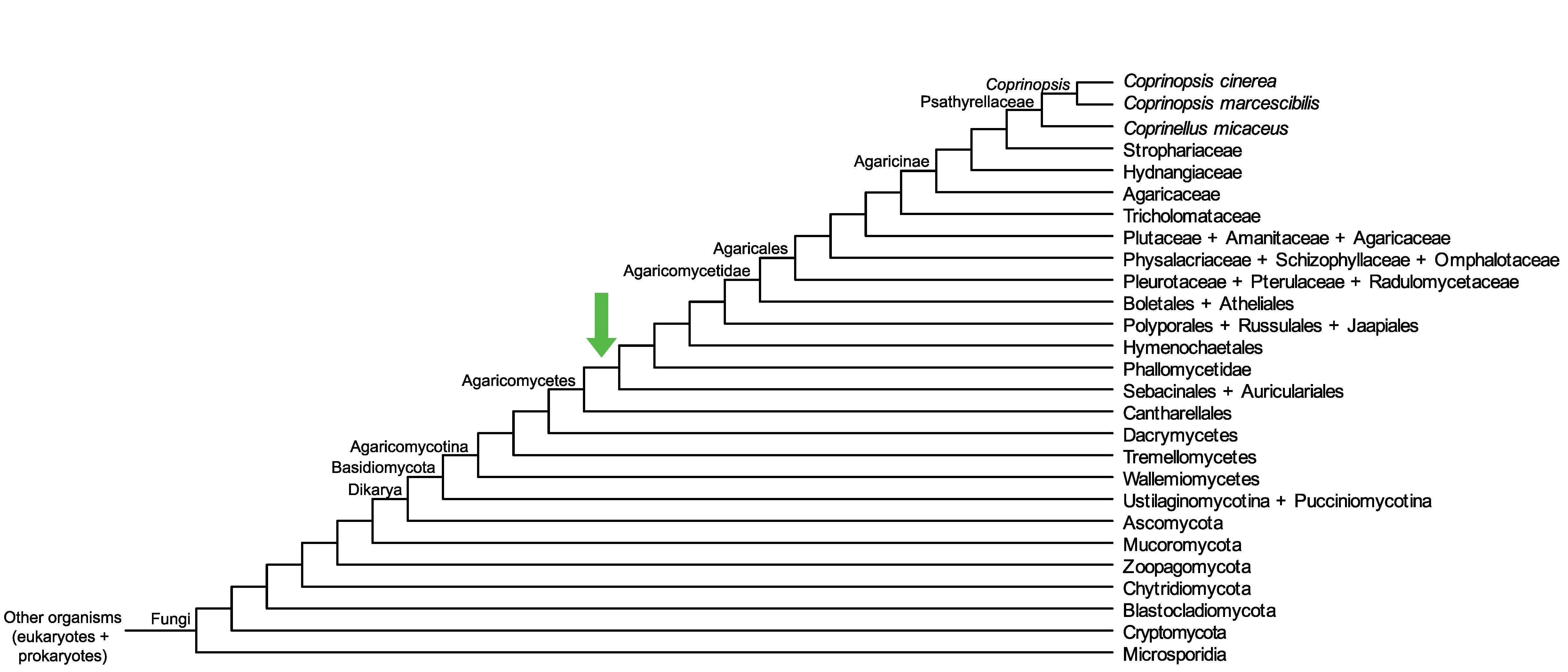
Conservation of CopciAB_374320 across fungi.
Arrow shows the origin of gene family containing CopciAB_374320.
Protein
| Sequence id | CopciAB_374320.T0 |
|---|---|
| Sequence |
>CopciAB_374320.T0 MSAVNEQKAVATNAIKVLTVVDWAQTSKGKPVYVQFTRQKPPPVEATRTWRGEVLPNPPPRPPSKLPPPACNELV VSTQNAIGQGRCGTIFAVDTMRMTNPKNPSFRLFHPRAPYLPDLVIKVAALDRAESLAHEAAMYEEMELLQGVSL PRCYGLFTANLPSDIVIPALGSEKLPPLTPSNPVQSNNRQVSILLLERVGDPLPLGDKLQEEGDLWDVFRDLARM GIEQTDIRYSNILQAPNSSGSFPSTVCPFHGTVHNYRIIDFDSARKTDLTLKRHYFNTLEYLGPILENMKQGVIV EPWDIKTKPDRPSAL |
| Length | 315 |
Coding
| Sequence id | CopciAB_374320.T0 |
|---|---|
| Sequence |
>CopciAB_374320.T0 ATGAGCGCCGTCAACGAGCAGAAAGCAGTGGCTACCAATGCCATCAAGGTCCTAACCGTCGTTGACTGGGCCCAA ACATCCAAAGGGAAACCTGTTTACGTGCAGTTCACCAGGCAGAAGCCTCCACCAGTTGAGGCCACACGTACATGG CGAGGAGAGGTGCTTCCAAACCCTCCTCCCCGTCCCCCATCCAAACTTCCTCCCCCGGCCTGCAACGAGCTCGTA GTCTCGACCCAAAATGCCATTGGTCAAGGTCGTTGTGGGACCATATTCGCTGTGGACACCATGCGAATGACCAAC CCCAAGAACCCGTCGTTTAGACTATTCCATCCCCGAGCTCCCTATCTGCCCGACCTGGTCATAAAAGTGGCCGCT CTCGATCGTGCTGAAAGTCTCGCCCACGAGGCTGCCATGTACGAGGAGATGGAGCTGCTCCAAGGCGTCTCTCTT CCTCGTTGCTATGGCTTGTTCACGGCCAACCTCCCCTCCGACATCGTGATCCCCGCATTGGGTTCTGAGAAGCTC CCGCCCTTGACACCCTCTAACCCCGTCCAATCCAATAACAGACAAGTGTCAATCCTTCTACTCGAGCGAGTCGGA GATCCATTGCCACTCGGCGACAAACTCCAGGAAGAAGGCGATCTTTGGGACGTATTCCGGGATCTCGCCCGCATG GGTATTGAACAAACCGACATACGGTATTCCAACATCCTCCAGGCGCCGAACTCGTCGGGCAGCTTCCCAAGCACC GTCTGCCCCTTCCACGGCACAGTCCACAATTACCGCATCATCGACTTCGATTCCGCAAGGAAGACTGACCTTACG TTAAAACGACACTACTTCAATACCCTCGAATACCTCGGTCCCATCCTTGAAAACATGAAGCAGGGCGTCATCGTC GAGCCATGGGACATCAAAACGAAACCCGACAGGCCAAGCGCTCTGTGA |
| Length | 948 |
Transcript
| Sequence id | CopciAB_374320.T0 |
|---|---|
| Sequence |
>CopciAB_374320.T0 ACAAATATCCCCCAAGCCCTCTTTTTTCTTCACACATTCAGAACTCTTCGTTATGAGCGCCGTCAACGAGCAGAA AGCAGTGGCTACCAATGCCATCAAGGTCCTAACCGTCGTTGACTGGGCCCAAACATCCAAAGGGAAACCTGTTTA CGTGCAGTTCACCAGGCAGAAGCCTCCACCAGTTGAGGCCACACGTACATGGCGAGGAGAGGTGCTTCCAAACCC TCCTCCCCGTCCCCCATCCAAACTTCCTCCCCCGGCCTGCAACGAGCTCGTAGTCTCGACCCAAAATGCCATTGG TCAAGGTCGTTGTGGGACCATATTCGCTGTGGACACCATGCGAATGACCAACCCCAAGAACCCGTCGTTTAGACT ATTCCATCCCCGAGCTCCCTATCTGCCCGACCTGGTCATAAAAGTGGCCGCTCTCGATCGTGCTGAAAGTCTCGC CCACGAGGCTGCCATGTACGAGGAGATGGAGCTGCTCCAAGGCGTCTCTCTTCCTCGTTGCTATGGCTTGTTCAC GGCCAACCTCCCCTCCGACATCGTGATCCCCGCATTGGGTTCTGAGAAGCTCCCGCCCTTGACACCCTCTAACCC CGTCCAATCCAATAACAGACAAGTGTCAATCCTTCTACTCGAGCGAGTCGGAGATCCATTGCCACTCGGCGACAA ACTCCAGGAAGAAGGCGATCTTTGGGACGTATTCCGGGATCTCGCCCGCATGGGTATTGAACAAACCGACATACG GTATTCCAACATCCTCCAGGCGCCGAACTCGTCGGGCAGCTTCCCAAGCACCGTCTGCCCCTTCCACGGCACAGT CCACAATTACCGCATCATCGACTTCGATTCCGCAAGGAAGACTGACCTTACGTTAAAACGACACTACTTCAATAC CCTCGAATACCTCGGTCCCATCCTTGAAAACATGAAGCAGGGCGTCATCGTCGAGCCATGGGACATCAAAACGAA ACCCGACAGGCCAAGCGCTCTGTGATACCCACCCCCTCCTGGACCCACTATCACACTCACAGCAACTCAACTTGC ATCACATGTACTACTAGTCTCGTCATGTCCCCCGCATGACCCTGCATTCAACATTGGACTTTGAATCATCACATT TTGTCACCCCATTATCGGTTTCTAAATCGAGCATGTCGTCGCTATTGTTTATACCTTGGATCTTACCTGCTATGT CTGTTTTCTGGCCTCAATGTGACCGCAACATTTTG |
| Length | 1235 |
Gene
| Sequence id | CopciAB_374320.T0 |
|---|---|
| Sequence |
>CopciAB_374320.T0 ACAAATATCCCCCAAGCCCTCTTTTTTCTTCACACATTCAGAACTCTTCGTTATGAGCGCCGTCAACGAGCAGAA AGCAGTGGTAAGTCGTCTGACCAGCTCTATATCAACGTTCGAGGACGCTCACACCTAGTTTCAGGCTACCAATGC CATCAAGGTCCTAACCGTCGTTGACTGGGCCCAAACATCCAAAGGGAAACCTGTTTACGTGCAGTTCACCAGGCA GAAGCCTCCACCAGTTGAGGCCACACGTACATGGCGAGGAGAGGTGCTTCCAAACCCTCCTCCCCGTCCCCCATC CAAACTTCCTCCCCCGGCCTGCAACGAGCTCGTAGTCTCGACCCAAAATGCCATTGGTCAAGGTCGTTGTGGGAC CATATTCGCTGTGGACACCATGCGAATGACCAACCCCAAGAACCCGTCGTTTAGACTATTCCATCCCCGAGCTCC CTATCTGCCCGACCTGGTCATAAAAGTGGCCGCTCTCGATCGTGCTGAAAGTCTCGCCCACGAGGCTGCCATGTA CGAGGAGATGGAGCTGCTCCAAGGCGTCTCTCTTCCTCGTTGCTATGGCTTGTTCACGGCCAACCTCCCCTCCGA CATCGTGATCCCCGCATTGGGTTCTGAGAAGCTCCCGCCCTTGACACCCTCTAACCCCGTCCAATCCAATAACAG ACAAGTGTCAATCCTTCTACTCGAGCGAGTCGGAGATCCATTGCCACTCGGCGACAAACTCCAGGAAGAGTAAGT CACTCCAACAGCTTGGCTCGAAGCAGGTGACCGATGAAACTGACATGCTCCTCACAGAGGCGATCTTTGGGACGT ATTCCGGGATCTCGCCCGCATGGGTATTGAACAAACCGACATACGGTATTCCAACATCCTCCAGGCGCCGAACTC GTCGGGCAGCTTCCCAAGCACCGTCTGCCCCTTCCACGGCACAGTCCACAATTACCGCATCATCGACTTCGATTC CGCAAGGAAGACTGACCTTACGTTAAAACGACACTACTTCAATACCCTCGAATACCTCGGTCCCATCCTTGAAAA CATGAAGCAGGGCGTCATCGTCGAGCCATGGGACATCAAAACGAAACCCGACAGGCCAAGCGCTCTGTGATACCC ACCCCCTCCTGGACCCACTATCACACTCACAGCAACTCAACTTGCATCACATGTACTACTAGTCTCGTCATGTCC CCCGCATGACCCTGCATTCAACATTGGACTTTGAATCATCACATTTTGTCACCCCATTATCGGTTTCTAAATCGA GCATGTCGTCGCTATTGTTTATACCTTGGATCTTACCTGCTATGTCTGTTTTCTGGCCTCAATGTGACCGCAACA TTTTG |
| Length | 1355 |