CopciAB_375402
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_375402 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 375402 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_1:3946082..3947616 | Strand | - |
| Gene length (nt) | 1535 | Transcript length (nt) | 1258 |
| CDS length (nt) | 858 | Protein length (aa) | 285 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_1938 | 36.1 | 6.413E-45 | 164 |
| Lentinula edodes B17 | Lened_B_1_1_4505 | 37.1 | 1.197E-43 | 160 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB27269 | 34.5 | 2.005E-41 | 154 |
| Agrocybe aegerita | Agrae_CAA7269732 | 30.5 | 5.603E-34 | 132 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_375402.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 14 | 271 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
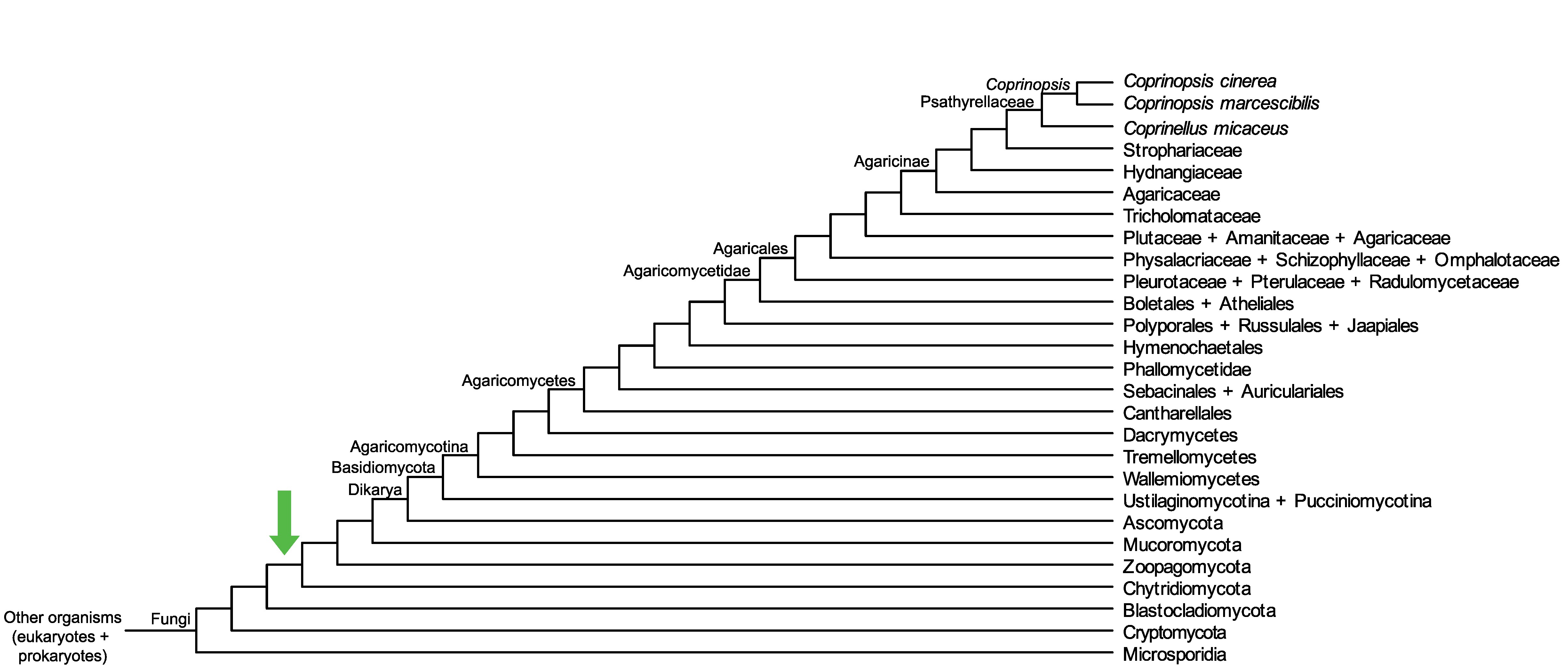
Conservation of CopciAB_375402 across fungi.
Arrow shows the origin of gene family containing CopciAB_375402.
Protein
| Sequence id | CopciAB_375402.T0 |
|---|---|
| Sequence |
>CopciAB_375402.T0 MPYRDFGPYTDVLDATHNCQKAVVKVFRIGKLSPHHRHVLTKRLENALTEWKRHHMHPNIARVTGMVLGFGDLPG IEMPLYSNGNAYRYLKAHPNVDKYRLICGIAEGLNHLHSQQPPIIHGMVCATNILISDSGAPLLTDIGLGTIPLE AVGVSPFQGDKYSRHLQKVRWLAPEIIDPENVGPPDISTSLNDCTPSTDVYSFGMTILELFTEKVPYHHVRSTPA VINEINNRRHPGRLYCTEVPDELWCILESCWSHHAKERPTTAVLRSRIGALGIHGSGRRA |
| Length | 285 |
Coding
| Sequence id | CopciAB_375402.T0 |
|---|---|
| Sequence |
>CopciAB_375402.T0 ATGCCGTATCGTGACTTCGGGCCCTACACAGACGTCCTCGACGCGACCCACAACTGTCAGAAAGCAGTTGTCAAA GTCTTTCGGATTGGAAAGCTCTCTCCTCACCACCGACATGTACTGACTAAGAGGCTAGAGAATGCGTTGACGGAG TGGAAGAGGCATCACATGCACCCTAACATTGCCAGAGTAACGGGAATGGTTCTCGGTTTCGGGGATCTTCCTGGT ATCGAAATGCCTCTGTACAGCAACGGGAATGCATACAGATATCTCAAGGCCCATCCAAATGTCGACAAGTACCGC TTGATCTGTGGGATTGCTGAGGGACTAAACCACCTGCATTCCCAACAACCACCCATCATACACGGCATGGTGTGC GCGACGAACATCTTAATCTCTGACTCGGGCGCCCCTCTGCTGACTGACATTGGTCTCGGAACAATTCCTCTAGAA GCTGTCGGAGTTTCGCCGTTCCAAGGTGATAAGTATTCTCGACATCTTCAGAAAGTTAGGTGGCTTGCCCCCGAG ATAATTGATCCAGAGAACGTGGGTCCCCCTGATATTAGTACGAGCTTAAACGATTGCACACCATCGACAGATGTC TACAGCTTTGGGATGACTATCCTGGAGCTATTCACCGAGAAGGTGCCATACCACCATGTCAGATCTACTCCTGCT GTAATCAACGAAATAAACAACCGCCGCCATCCAGGCCGATTGTACTGCACCGAGGTGCCTGACGAGCTATGGTGC ATCCTTGAAAGCTGCTGGAGTCATCACGCGAAGGAGCGACCAACCACTGCGGTCCTGCGCTCGCGGATAGGGGCT CTCGGCATACACGGCAGCGGCCGGAGGGCTTAG |
| Length | 858 |
Transcript
| Sequence id | CopciAB_375402.T0 |
|---|---|
| Sequence |
>CopciAB_375402.T0 AGAGTCTACGACCATTCTACCTCGACGACGGGCGTCTAACAAGGCGCCTTCCATATGATTCGAACGTCGTCCTAT ACTTGTGTGTCTGGGACTGCGGAAGCTGTTACTAGGAGGGCTATATAGCCAGCTCCTGCTCACCCTTCCTTGCTG TAAACACCCACCCACCCATACATGCACACATTCACAATTATCAGGTCGACTTCCACCACGATATCACTACAAGCG CTTACCGACTCTCGGATCGAGGGTTGAAATTACAGCTGGACATGCCGTATCGTGACTTCGGGCCCTACACAGACG TCCTCGACGCGACCCACAACTGTCAGAAAGCAGTTGTCAAAGTCTTTCGGATTGGAAAGCTCTCTCCTCACCACC GACATGTACTGACTAAGAGGCTAGAGAATGCGTTGACGGAGTGGAAGAGGCATCACATGCACCCTAACATTGCCA GAGTAACGGGAATGGTTCTCGGTTTCGGGGATCTTCCTGGTATCGAAATGCCTCTGTACAGCAACGGGAATGCAT ACAGATATCTCAAGGCCCATCCAAATGTCGACAAGTACCGCTTGATCTGTGGGATTGCTGAGGGACTAAACCACC TGCATTCCCAACAACCACCCATCATACACGGCATGGTGTGCGCGACGAACATCTTAATCTCTGACTCGGGCGCCC CTCTGCTGACTGACATTGGTCTCGGAACAATTCCTCTAGAAGCTGTCGGAGTTTCGCCGTTCCAAGGTGATAAGT ATTCTCGACATCTTCAGAAAGTTAGGTGGCTTGCCCCCGAGATAATTGATCCAGAGAACGTGGGTCCCCCTGATA TTAGTACGAGCTTAAACGATTGCACACCATCGACAGATGTCTACAGCTTTGGGATGACTATCCTGGAGCTATTCA CCGAGAAGGTGCCATACCACCATGTCAGATCTACTCCTGCTGTAATCAACGAAATAAACAACCGCCGCCATCCAG GCCGATTGTACTGCACCGAGGTGCCTGACGAGCTATGGTGCATCCTTGAAAGCTGCTGGAGTCATCACGCGAAGG AGCGACCAACCACTGCGGTCCTGCGCTCGCGGATAGGGGCTCTCGGCATACACGGCAGCGGCCGGAGGGCTTAGT AACGTGTGCCGTAGGCCCGCGGTAGGGACAAAAACAGCCCGAAAGAGGAACTCCGACTCGGAGACTGAATTTCTG GTATATAAACCCTGACGTCGTTGTCTACGATCGCAATGCAATTTTGCTTTACACCATC |
| Length | 1258 |
Gene
| Sequence id | CopciAB_375402.T0 |
|---|---|
| Sequence |
>CopciAB_375402.T0 AGAGTCTACGACCATTCTACCTCGACGACGGGCGTCTAACAAGGCGCCTTCCATATGATTCGAACGTCGTCCTAT ACTTGTGTGTCTGGGACTGCGGAAGCTGTTACTAGGAGGGCTATATAGCCAGCTCCTGCTCACCCTTCCTTGCTG TAAACACCCACCCACCCATACATGCACACATTCACAATTATCAGGTATATCCTTCAACGCATCCAATATCTCAAG CTAAATGTTGTCTTAGGTCGACTTCCACCACGATATCACTACAAGCGCTTACCGACTCTCGGATCGAGGGTTGAA ATTACAGCTGGACATGCCGTATCGTGACTTCGGGCCCTACACAGACGTCCTCGACGCGACCCACAACTGTCAGAA AGCAGTTGTCAAAGTCTTTCGGATTGGAAAGCTCTCTCCTCACCACCGACATGTACTGACTAAGGTATGTGTATT TCGGTCAAAAAGAATCCATCGAGCGGTTTGGCTATATCGTAACATATGCAGAGGCTAGAGAATGCGTTGACGGAG TGGAAGAGGCATCACATGCACCCTAACATTGCCAGAGTAACGGGAATGGTTCTCGGTTTCGGGGATCTTCCTGGT ATCGAAATGCCTCTGTACAGCAACGGGAATGCATACAGATATCTCAAGGCCCATCCAAATGTCGACAAGTACCGC TTGGTAGGTGGTCAATACGTGAATTGGGAGTCGCTGTGCATAATCTGACGCCCACGGAAGATCTGTGGGATTGCT GAGGGACTAAACCACCTGCATTCCCAACAACCACCCATCATACACGGCATGGTGTGCGCGGTGAGTATGTTTCAC TCACTTTCATTCTAGCCCCTGACAGATACTCCTCAGACGAACATCTTAATCTCTGACTCGGGCGCCCCTCTGCTG ACTGACATTGGTCTCGGAACAATTCCTCTAGAAGCTGTCGGAGTTTCGCCGTTCCAAGGTGATAAGTATTCTCGA CATCTTCAGAAAGTTAGGTGGCTTGCCCCCGAGATAATTGATCCAGAGAACGTGGGTCCCCCTGATATTAGTACG AGCTTAAACGATTGCACACCATCGACAGATGTCTACAGCTTTGGGATGACTATCCTGGAGGTCCGTCTACAACGT GATTGATAAGTATTAAGCGCTGGTCAACTAACACCCGATCCACAGCTATTCACCGAGAAGGTGCCATACCACCAT GTCAGATCTACTCCTGCTGTAATCAACGAAATAAACAACCGCCGCCATCCAGGCCGATTGTACTGCACCGAGGTG CCTGACGAGCTATGGTGCATCCTTGAAAGCTGCTGGAGTCATCACGCGAAGGAGCGACCAACCACTGCGGTCCTG CGCTCGCGGATAGGGGCTCTCGGCATACACGGCAGCGGCCGGAGGGCTTAGTAACGTGTGCCGTAGGCCCGCGGT AGGGACAAAAACAGCCCGAAAGAGGAACTCCGACTCGGAGACTGAATTTCTGGTATATAAACCCTGACGTCGTTG TCTACGATCGCAATGCAATTTTGCTTTACACCATC |
| Length | 1535 |