CopciAB_378005
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_378005 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 378005 |
| Uniprot id | Functional description | ||
| Location | scaffold_2:755671..759206 | Strand | - |
| Gene length (nt) | 3536 | Transcript length (nt) | 3175 |
| CDS length (nt) | 2745 | Protein length (aa) | 914 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| 0 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_378005.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 279 | 736 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR040976 | Fungal-type protein kinase |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
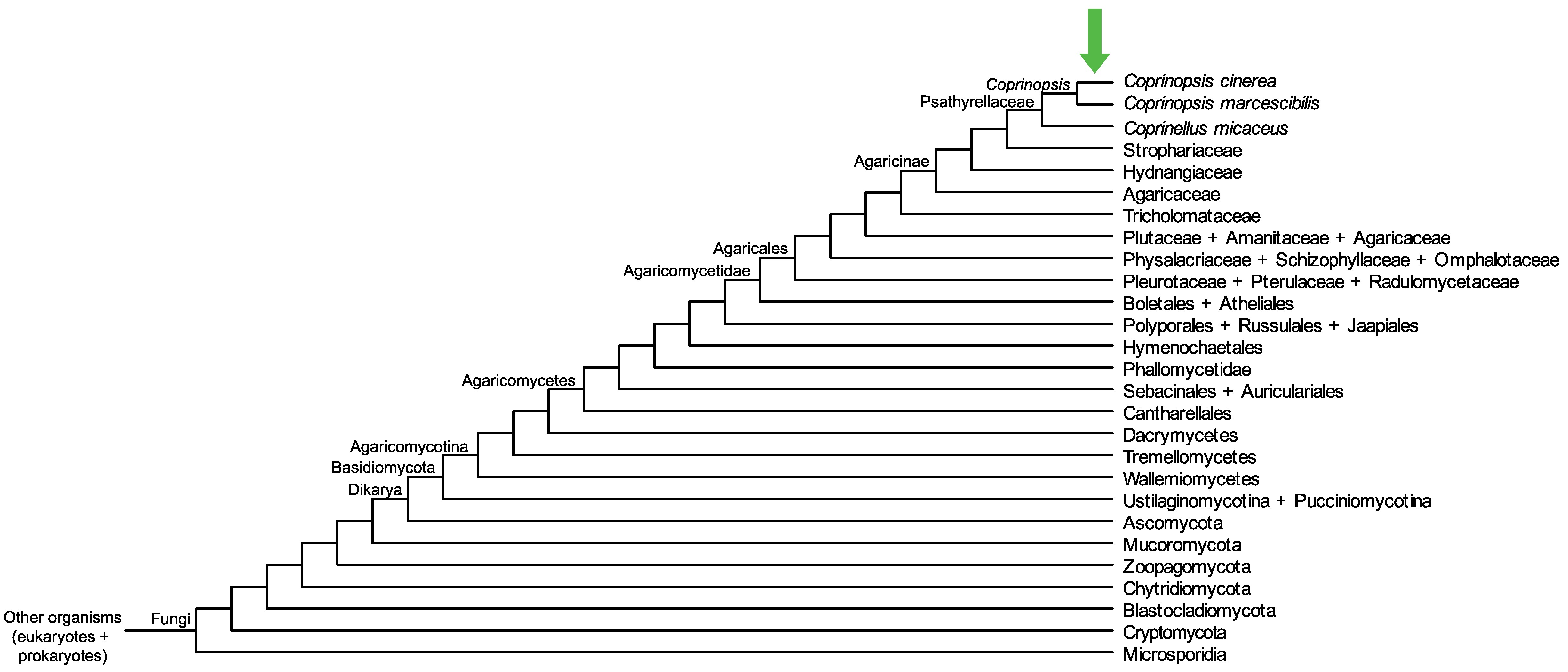
Conservation of CopciAB_378005 across fungi.
Arrow shows the origin of gene family containing CopciAB_378005.
Protein
| Sequence id | CopciAB_378005.T0 |
|---|---|
| Sequence |
>CopciAB_378005.T0 MKASTCPITPPRNPAGEPVRGQFTAPALGTSPRSAPNRPDVDSTTTNMRMDIGRIMNFEMYICTYNQFKGSYLPK CDEETLEFVLECLKALGESPFSQILVPREEDERVDARSAGENDKTVSLDDKETEEVSERNEAEPAVLLAEGNEGD IGDCESEDVEQGYISDEEGEGDQGDERKGNSKPEIYSHVFRDFKNKPSLLKKGANANQNSKTSSSDKTKNPNIHE DKVFNPFGEIGLRILAALDKKGVKRNGYHLRMCPHTTIYSNVPGCNQEIDACLTKNKSKSLDIADILVPWELKLS YTIAQVFQNRRQLASQVNHTMNDDPRRKFMFGITIEDDRVSLWFFSRSHSVKAKSFSMVERPDLLVEVLVSLLCA TDEQLGLDPLVHLTEDRRYVYEFPPTSTRPSPIYYRITETVSEFRWLRLICRATRIWKVIQVETPAHTEGIEGAK ERILKDVCSVAEFLTESEIQHQLFGDIEAFAKDKEWRQREILQEFVEEDMATLGQALEGDFRRFFSCIIDEHIGE VTPPLCAGAWVATTPVFIEPQTKSQEHSKNRAAPGHRADPRPEVLQDEHVIVEEIDPSRAFVPRRQCRYIYEEVC TPLNDIPTLGEAVDIIQQCLIALRLMFCAGWVHRDISYGNILAYRPSPETQWHVKLSDLEYAKRFPSDAVASTIP KTGTPYFMACEILTGKLITSAAAREPTLKRGGIRRNKVVNISIVFHTYQHDLESIWWILFWKITMRIKQVLPRAV PEQYFKNVISRAHASYRNDLFVSRQELAEDNDLCKSLPPTLKDSDFLYSLDALRNEFEVEYSNRNIAGKQREVGT YSVIISKAFWKFFEGIKDTRGEWGSLPLIVDADLRRAKMLLQKAPTATKRQFSELHPGLKGNMPAKSSAEPVKGV MTATHGSGSKRKCL |
| Length | 914 |
Coding
| Sequence id | CopciAB_378005.T0 |
|---|---|
| Sequence |
>CopciAB_378005.T0 ATGAAAGCCTCCACGTGTCCGATTACGCCTCCTCGCAATCCGGCGGGAGAGCCTGTCCGCGGACAGTTCACCGCT CCTGCACTTGGTACTTCACCACGCTCAGCCCCAAACCGACCCGACGTAGACAGTACCACCACAAACATGCGTATG GACATCGGTCGCATCATGAACTTCGAAATGTACATCTGCACGTACAATCAGTTCAAAGGAAGCTATCTACCGAAG TGTGATGAGGAGACGTTGGAATTTGTTTTGGAGTGTCTGAAGGCACTGGGAGAGTCACCTTTCAGCCAAATTCTC GTCCCTCGCGAAGAAGATGAAAGAGTTGATGCCAGATCTGCCGGGGAGAATGATAAAACGGTATCACTTGATGAT AAGGAGACAGAAGAGGTTAGTGAGCGCAACGAAGCGGAGCCTGCTGTTCTGCTGGCAGAAGGCAATGAAGGCGAC ATAGGAGATTGTGAGAGCGAGGATGTTGAGCAGGGGTATATTTCCGACGAAGAAGGCGAGGGTGACCAAGGTGAT GAGAGGAAAGGTAACTCGAAGCCGGAGATCTACTCTCACGTCTTCCGGGACTTCAAGAACAAGCCGTCCTTGTTG AAGAAGGGAGCAAACGCCAATCAGAACTCGAAGACCAGTTCGTCTGACAAGACAAAGAATCCCAACATTCACGAG GATAAGGTCTTCAACCCATTCGGCGAGATTGGACTCAGGATCCTAGCAGCCTTGGACAAGAAAGGTGTCAAGAGA AATGGGTATCACCTTCGCATGTGTCCACATACAACAATCTATTCCAACGTCCCCGGATGCAATCAAGAAATTGAC GCTTGCTTGACGAAGAACAAATCGAAATCCCTTGATATTGCCGACATTCTTGTTCCTTGGGAACTCAAGTTGTCC TACACCATTGCTCAGGTCTTTCAGAATCGGCGACAGCTTGCGTCTCAAGTTAATCACACGATGAACGATGATCCT CGACGGAAATTCATGTTCGGAATTACCATTGAAGATGACCGCGTATCCCTCTGGTTTTTCTCTCGGTCACATTCC GTGAAAGCGAAATCGTTTAGTATGGTTGAACGACCTGATCTTCTAGTAGAGGTTCTTGTCTCGCTCTTGTGCGCA ACAGACGAACAGCTTGGATTGGACCCTCTTGTTCATTTGACCGAGGACCGACGATACGTCTACGAATTTCCTCCG ACCAGTACGCGGCCCAGTCCCATTTACTATCGCATTACGGAAACAGTCTCTGAATTCCGATGGCTGAGACTCATC TGCCGTGCCACACGCATCTGGAAGGTGATCCAGGTGGAGACGCCGGCCCATACCGAGGGCATTGAAGGGGCAAAG GAAAGAATCCTCAAGGACGTTTGCTCTGTGGCTGAATTCCTCACGGAATCCGAGATTCAGCACCAACTGTTTGGA GACATCGAAGCGTTTGCAAAGGACAAGGAGTGGCGTCAGAGGGAAATACTTCAGGAGTTCGTCGAGGAAGATATG GCCACCCTTGGTCAGGCATTGGAAGGCGATTTTAGGAGGTTCTTCTCCTGTATCATCGATGAACACATCGGAGAG GTTACCCCGCCACTTTGTGCTGGCGCATGGGTTGCCACAACCCCAGTGTTTATCGAGCCCCAGACCAAATCCCAA GAGCACAGCAAGAATCGGGCGGCCCCAGGTCATCGGGCAGACCCTAGACCCGAGGTCCTGCAGGACGAGCATGTT ATTGTTGAGGAAATAGACCCTTCCCGGGCATTTGTGCCCAGACGCCAGTGTCGCTACATTTATGAGGAGGTTTGT ACACCGTTAAACGATATCCCAACCCTGGGGGAAGCCGTCGACATCATCCAACAATGCCTTATCGCTCTTCGCCTC ATGTTCTGTGCAGGATGGGTCCATCGAGACATAAGCTACGGAAATATTTTGGCGTATCGCCCCTCGCCGGAGACA CAGTGGCACGTCAAGCTATCCGACCTTGAGTACGCGAAGCGATTTCCTTCCGATGCTGTCGCTAGCACCATCCCC AAGACGGGCACACCGTACTTCATGGCATGCGAAATCTTGACTGGGAAGTTGATCACCAGCGCCGCCGCAAGGGAG CCAACTTTAAAGCGGGGCGGTATCAGACGCAACAAAGTTGTGAATATCAGCATTGTCTTCCACACCTACCAGCAC GATCTGGAATCCATCTGGTGGATTTTATTTTGGAAGATAACGATGCGAATCAAGCAAGTTCTTCCAAGGGCTGTC CCAGAGCAGTACTTCAAGAACGTTATCAGTCGCGCCCATGCATCATACCGTAACGACCTTTTCGTTTCTCGCCAG GAACTTGCAGAGGACAATGACCTTTGCAAATCGCTTCCACCGACATTAAAGGACTCTGACTTTCTCTATTCCCTG GACGCCCTTAGAAACGAGTTCGAAGTGGAGTATTCCAATCGCAACATAGCCGGCAAACAGCGCGAGGTCGGCACA TATTCCGTCATTATCAGCAAGGCGTTTTGGAAGTTCTTTGAAGGCATCAAGGATACCAGGGGTGAATGGGGGAGC CTCCCATTGATAGTTGACGCGGACCTACGTCGAGCTAAAATGCTTCTACAGAAGGCACCCACAGCGACGAAACGT CAATTCAGCGAGCTTCACCCAGGTCTTAAGGGTAATATGCCTGCTAAATCCAGCGCAGAACCTGTAAAAGGCGTG ATGACAGCGACACACGGTTCGGGTTCAAAGCGGAAGTGTCTCTGA |
| Length | 2745 |
Transcript
| Sequence id | CopciAB_378005.T0 |
|---|---|
| Sequence |
>CopciAB_378005.T0 CCATCACTCGACGCGTCTTGACTCTCAACCACATTCTCGTGTAAGAAACTCTACATAAGTGTCGTTCATTTCAGT TGCTCGAGCTTCGGGGCTACCATGAAAGCCTCCACGTGTCCGATTACGCCTCCTCGCAATCCGGCGGGAGAGCCT GTCCGCGGACAGTTCACCGCTCCTGCACTTGGTACTTCACCACGCTCAGCCCCAAACCGACCCGACGTAGACAGT ACCACCACAAACATGCGTATGGACATCGGTCGCATCATGAACTTCGAAATGTACATCTGCACGTACAATCAGTTC AAAGGAAGCTATCTACCGAAGTGTGATGAGGAGACGTTGGAATTTGTTTTGGAGTGTCTGAAGGCACTGGGAGAG TCACCTTTCAGCCAAATTCTCGTCCCTCGCGAAGAAGATGAAAGAGTTGATGCCAGATCTGCCGGGGAGAATGAT AAAACGGTATCACTTGATGATAAGGAGACAGAAGAGGTTAGTGAGCGCAACGAAGCGGAGCCTGCTGTTCTGCTG GCAGAAGGCAATGAAGGCGACATAGGAGATTGTGAGAGCGAGGATGTTGAGCAGGGGTATATTTCCGACGAAGAA GGCGAGGGTGACCAAGGTGATGAGAGGAAAGGTAACTCGAAGCCGGAGATCTACTCTCACGTCTTCCGGGACTTC AAGAACAAGCCGTCCTTGTTGAAGAAGGGAGCAAACGCCAATCAGAACTCGAAGACCAGTTCGTCTGACAAGACA AAGAATCCCAACATTCACGAGGATAAGGTCTTCAACCCATTCGGCGAGATTGGACTCAGGATCCTAGCAGCCTTG GACAAGAAAGGTGTCAAGAGAAATGGGTATCACCTTCGCATGTGTCCACATACAACAATCTATTCCAACGTCCCC GGATGCAATCAAGAAATTGACGCTTGCTTGACGAAGAACAAATCGAAATCCCTTGATATTGCCGACATTCTTGTT CCTTGGGAACTCAAGTTGTCCTACACCATTGCTCAGGTCTTTCAGAATCGGCGACAGCTTGCGTCTCAAGTTAAT CACACGATGAACGATGATCCTCGACGGAAATTCATGTTCGGAATTACCATTGAAGATGACCGCGTATCCCTCTGG TTTTTCTCTCGGTCACATTCCGTGAAAGCGAAATCGTTTAGTATGGTTGAACGACCTGATCTTCTAGTAGAGGTT CTTGTCTCGCTCTTGTGCGCAACAGACGAACAGCTTGGATTGGACCCTCTTGTTCATTTGACCGAGGACCGACGA TACGTCTACGAATTTCCTCCGACCAGTACGCGGCCCAGTCCCATTTACTATCGCATTACGGAAACAGTCTCTGAA TTCCGATGGCTGAGACTCATCTGCCGTGCCACACGCATCTGGAAGGTGATCCAGGTGGAGACGCCGGCCCATACC GAGGGCATTGAAGGGGCAAAGGAAAGAATCCTCAAGGACGTTTGCTCTGTGGCTGAATTCCTCACGGAATCCGAG ATTCAGCACCAACTGTTTGGAGACATCGAAGCGTTTGCAAAGGACAAGGAGTGGCGTCAGAGGGAAATACTTCAG GAGTTCGTCGAGGAAGATATGGCCACCCTTGGTCAGGCATTGGAAGGCGATTTTAGGAGGTTCTTCTCCTGTATC ATCGATGAACACATCGGAGAGGTTACCCCGCCACTTTGTGCTGGCGCATGGGTTGCCACAACCCCAGTGTTTATC GAGCCCCAGACCAAATCCCAAGAGCACAGCAAGAATCGGGCGGCCCCAGGTCATCGGGCAGACCCTAGACCCGAG GTCCTGCAGGACGAGCATGTTATTGTTGAGGAAATAGACCCTTCCCGGGCATTTGTGCCCAGACGCCAGTGTCGC TACATTTATGAGGAGGTTTGTACACCGTTAAACGATATCCCAACCCTGGGGGAAGCCGTCGACATCATCCAACAA TGCCTTATCGCTCTTCGCCTCATGTTCTGTGCAGGATGGGTCCATCGAGACATAAGCTACGGAAATATTTTGGCG TATCGCCCCTCGCCGGAGACACAGTGGCACGTCAAGCTATCCGACCTTGAGTACGCGAAGCGATTTCCTTCCGAT GCTGTCGCTAGCACCATCCCCAAGACGGGCACACCGTACTTCATGGCATGCGAAATCTTGACTGGGAAGTTGATC ACCAGCGCCGCCGCAAGGGAGCCAACTTTAAAGCGGGGCGGTATCAGACGCAACAAAGTTGTGAATATCAGCATT GTCTTCCACACCTACCAGCACGATCTGGAATCCATCTGGTGGATTTTATTTTGGAAGATAACGATGCGAATCAAG CAAGTTCTTCCAAGGGCTGTCCCAGAGCAGTACTTCAAGAACGTTATCAGTCGCGCCCATGCATCATACCGTAAC GACCTTTTCGTTTCTCGCCAGGAACTTGCAGAGGACAATGACCTTTGCAAATCGCTTCCACCGACATTAAAGGAC TCTGACTTTCTCTATTCCCTGGACGCCCTTAGAAACGAGTTCGAAGTGGAGTATTCCAATCGCAACATAGCCGGC AAACAGCGCGAGGTCGGCACATATTCCGTCATTATCAGCAAGGCGTTTTGGAAGTTCTTTGAAGGCATCAAGGAT ACCAGGGGTGAATGGGGGAGCCTCCCATTGATAGTTGACGCGGACCTACGTCGAGCTAAAATGCTTCTACAGAAG GCACCCACAGCGACGAAACGTCAATTCAGCGAGCTTCACCCAGGTCTTAAGGGTAATATGCCTGCTAAATCCAGC GCAGAACCTGTAAAAGGCGTGATGACAGCGACACACGGTTCGGGTTCAAAGCGGAAGTGTCTCTGACGACGGATA AAGGAGCGGCCTCGAGAGCCAATACGAAGAGCACTGCCAAAATCAGTGACGAGGGCGACGAGACAAGGCCATCGA AGAAGGCTCGACGGTGAATATCTTCTTCAAGCACGGTTGAGGGATGCATGATGTCAGTGAGGGTTCGCCAGTCGT TGCCAGTTCATGCATTGGTTGGTTATTCAGCGCATGGAATAGCTTAGGTCTGGGACTGGGACAAGCTCGGACTCT TCTCGTATATATCCTTTATCCTACTTGAGTAGTGTTGTACTCGGGCACTGTGGTCATTTTACATAAGAAGTCCAT TGCGAATGCTACATGTATGAGCGTG |
| Length | 3175 |
Gene
| Sequence id | CopciAB_378005.T0 |
|---|---|
| Sequence |
>CopciAB_378005.T0 CCATCACTCGACGCGTCTTGACTCTCAACCACATTCTCGTGTAAGAAACTCTACATAAGTGTCGTTCATTTCAGT TGCTCGAGCTTCGGGGCTACCATGAAAGCCTCCACGTGTCCGATGTAAGTCATTTCTGTGTTCATACCAGCTCGA AGTCTCCCGCTTGCCACTTCTCGATGCTCACAGTATTCTCCTAGTACGCCTCCTCGCAATCCGGCGGGAGAGCCT GTCCGCGGACAGTTCACCGCTCCTGCACTTGGTACTTCACCACGCTCAGCCCCAAACCGACCCGACGTAGACAGT ACCACCACAAACATGCGTATGGACATCGGTCGCATCATGAACTTCGAAATGTACATCTGCACGTACAATCAGTTC AAAGGAAGCTATCTACCGAAGTGTGATGAGGAGACGTTGGAATTTGTTTTGGAGTGTCTGAAGGCACTGGGAGAG TCACCTTTCAGCCAAATTCTCGTCCCTCGCGAAGAAGATGAAAGAGTTGATGCCAGATCTGCCGGGGAGAATGAT AAAACGGTATCACTTGATGATAAGGAGACAGAAGAGGTTAGTGAGCGCAACGAAGCGGAGCCTGCTGTTCTGCTG GCAGAAGGCAATGAAGGCGACATAGGAGATTGTGAGAGCGAGGATGTTGAGCAGGGGTATATTTCCGACGAAGAA GGCGAGGGTGACCAAGGTGATGAGAGGAAAGGTAACTCGAAGCCGGAGATCTACTCTCACGTCTTCCGGGACTTC AAGAACAAGCCGTCCTTGTTGAAGAAGGGAGCAAACGCCAATCAGAACTCGAAGACCAGTTCGTCTGACAAGACA AAGAATCCCAACATTCACGAGGATAAGGTCTTCAACCCATTCGGCGAGATTGGACTCAGGATCCTAGCAGCCTTG GACAAGAAAGGTGTCAAGAGAAATGGGTATCACCTTCGCATGTGTCCACATACAACAATCTATTCCAACGTCCCC GGATGCAATCAAGAAATTGACGCTTGCTTGACGAAGAACAAATCGAAATCCCTTGATATTGCCGACATTCTTGTT CCTTGGGAACTCAAGTTGTCCTACACCATTGCTCAGGTCTTTCAGGTGAGTGTAGTCCCTTTGGCCTTTCAGCTG TATCGGCAAGCTCAAAATATTTTCTAGAATCGGCGACAGCTTGCGTCTCAAGTTAATCACACGATGAACGATGAT CCTCGACGGAAATTCATGTTCGGAGTAAGTGTGTACCTCATCATGGTCTGACTTGTCCCTGATCTTACGGTGCCC CGTCCCAGATTACCATTGAAGATGACCGCGTATCCCTCTGGTTTTTCTCTCGGTCACATTCCGTGAAAGCGAAAT CGTTTAGTATGGTTGAAGTGCGTTTCCGTAATTCATTTGGTCTCAGCCTTGGCTAATGGTATTTCAGCGACCTGA TCTTCTAGTAGAGGTTCTTGTCTCGCTCTTGTGCGCAACAGACGAACAGCTTGGATTGGACCCTCTTGTTCATTT GACCGAGGACCGACGATACGTCTACGAATTTCCTCCGACCAGTACGCGGCCCAGTCCCATTTACTATCGCATTAC GGAAACAGTCTCTGAATTCCGATGGCTGAGACTCATCTGCCGTGCCACACGCATCTGGAAGGTGATCCAGGTGGA GACGCCGGCCCATACCGAGGGCATTGAAGGGGCAAAGGAAAGAATCCTCAAGGACGTTTGCTCTGTGGCTGAATT CCTCACGGAATCCGAGATTCAGCACCAACTGTTTGGAGACATCGAAGCGTTTGCAAAGGACAAGGAGTGGCGTCA GAGGGAAATACTTCAGGAGTTCGTCGAGGAAGATATGGCCACCCTTGGTCAGGCATTGGAAGGCGATTTTAGGAG GTTCTTCTCCTGTATCATCGATGAACACATCGGAGAGGTTACCCCGCCACTTTGTGCTGGCGCATGGGTTGCCAC AACCCCAGTGTTTATCGAGCCCCAGACCAAATCCCAAGAGCACAGCAAGAATCGGGCGGCCCCAGGTCATCGGGC AGACCCTAGACCCGAGGTCCTGCAGGACGAGCATGTTATTGTTGAGGAAATAGACCCTTCCCGGGCATTTGTGCC CAGACGCCAGTGTCGCTACATTTATGAGGAGGTTTGTACACCGTTAAACGATATCCCAACCCTGGGGGAAGCCGT CGACATCATCCAACAATGCCTTATCGGTATGTGGTCCAGTTCTGCATTAACCCCAGTCTGTCTTAGCTCACTTTT GGCTTTAGCTCTTCGCCTCATGTTCTGTGCAGGATGGGTCCATCGAGACATAAGCTACGGAAATATTTTGGCGTA TCGCCCCTCGCCGGAGACACAGTGGCACGTCAAGCTATCCGACCTTGAGTACGCGAAGCGATTTCCTTCCGATGC TGTCGCTAGCACCATCCCCAAGACGGTTTGTCTCACATTCAATCTACTGGTGACCAACCCCGGGCCGTCTTCTGA CAACCTTGCACAGGGCACACCGTACTTCATGGCATGCGAAATCTTGACTGGGAAGTTGATCACCAGCGCCGCCGC AAGGGAGCCAACTTTAAAGCGGGGCGGTATCAGACGCAACAAAGTTGTGAATATCAGCATTGTCTTCCACACCTA CCAGCACGATCTGGAATCCATCTGGTGGATTTTATTTTGGAAGATAACGATGCGAATCAAGCAAGTTCTTCCAAG GGCTGTCCCAGAGCAGTACTTCAAGAACGTTATCAGTCGCGCCCATGCATCATACCGTAACGACCTTTTCGTTTC TCGCCAGGAACTTGCAGAGGACAATGACCTTTGCAAATCGCTTCCACCGACATTAAAGGACTCTGACTTTCTCTA TTCCCTGGACGCCCTTAGAAACGAGTTCGAAGTGGAGTATTCCAATCGCAACATAGCCGGCAAACAGCGCGAGGT CGGCACATATTCCGTCATTATCAGCAAGGCGTTTTGGAAGTTCTTTGAAGGCATCAAGGATACCAGGGGTGAATG GGGGAGCCTCCCATTGATAGTTGACGCGGACCTACGTCGAGCTAAAATGCTTCTACAGAAGGCACCCACAGCGAC GAAACGTCAATTCAGCGAGCTTCACCCAGGTCTTAAGGGTAATATGCCTGCTAAATCCAGCGCAGAACCTGTAAA AGGCGTGATGACAGCGACACACGGTTCGGGTTCAAAGCGGAAGTGTCTCTGACGACGGATAAAGGAGCGGCCTCG AGAGCCAATACGAAGAGCACTGCCAAAATCAGTGACGAGGGCGACGAGACAAGGCCATCGAAGAAGGCTCGACGG TGAATATCTTCTTCAAGCACGGTTGAGGGATGCATGATGTCAGTGAGGGTTCGCCAGTCGTTGCCAGTTCATGCA TTGGTTGGTTATTCAGCGCATGGAATAGCTTAGGTCTGGGACTGGGACAAGCTCGGACTCTTCTCGTATATATCC TTTATCCTACTTGAGTAGTGTTGTACTCGGGCACTGTGGTCATTTTACATAAGAAGTCCATTGCGAATGCTACAT GTATGAGCGTG |
| Length | 3536 |