CopciAB_379729
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_379729 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 379729 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_6:958312..961600 | Strand | + |
| Gene length (nt) | 3289 | Transcript length (nt) | 2556 |
| CDS length (nt) | 2166 | Protein length (aa) | 721 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB23713 | 66.2 | 1.165E-291 | 919 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1441723 | 62 | 6.939E-274 | 850 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_9376 | 62.5 | 3.645E-272 | 845 |
| Lentinula edodes NBRC 111202 | Lenedo1_1270884 | 62.5 | 4.339E-270 | 839 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_54872 | 60.6 | 1.938E-267 | 831 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_223047 | 60.7 | 2.02E-266 | 828 |
| Lentinula edodes B17 | Lened_B_1_1_5156 | 61.6 | 5.198E-265 | 824 |
| Agrocybe aegerita | Agrae_CAA7267420 | 63.1 | 1.52E-264 | 823 |
| Schizophyllum commune H4-8 | Schco3_2617727 | 59.8 | 4.803E-251 | 784 |
| Grifola frondosa | Grifr_OBZ73830 | 50 | 8.692E-221 | 696 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_128991 | 57.7 | 1.924E-220 | 695 |
| Flammulina velutipes | Flave_chr07AA00633 | 54.1 | 6.593E-204 | 647 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1043792 | 80 | 1.111E-195 | 623 |
| Pleurotus ostreatus PC9 | PleosPC9_1_17918 | 80 | 1.087E-195 | 623 |
| Auricularia subglabra | Aurde3_1_1235015 | 44.3 | 3.106E-167 | 541 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_379729.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd07834 | STKc_MAPK | - | 96 | 425 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 98 | 387 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR003527 | Mitogen-activated protein (MAP) kinase, conserved site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004707 | MAP kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
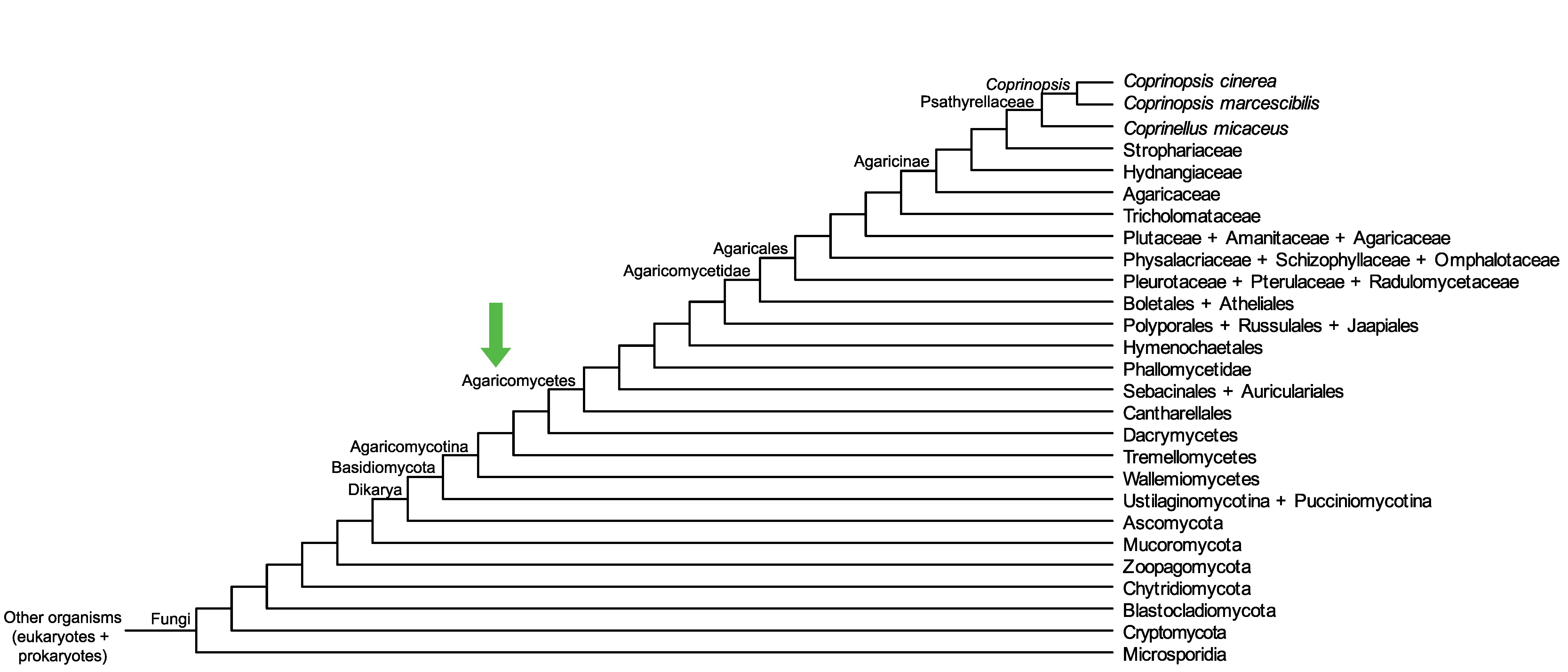
Conservation of CopciAB_379729 across fungi.
Arrow shows the origin of gene family containing CopciAB_379729.
Protein
| Sequence id | CopciAB_379729.T0 |
|---|---|
| Sequence |
>CopciAB_379729.T0 MSRVRGGHSPARSPSRPHRRRSTRGRSNGDENGNGDGSNDNNKSSSPVPSPGSWNVDRGRSRVRRPSNPFSEDNL RRRGYHSLHSSFGKVFHVEKRWKLVREMGSGAYGVVISAADEISGETVAIKMVTRVFEKIQLAKRALREITLLRH FANHENITGLIDVDAVSPDFNEIYIFMEPMEADLHQIIKSGQVLTNEHVQYFLYQILRGMKFIHSAGVIHRDLKP GNLLVNADCELKICDFGLSRGFDAVPDENATRLTEYVATRWYRAPEIMLAFPRYNTAIDVWSIGCILGELLLGKP LFKGKDYVDQLNKILDILGSPDETIIQKIGSDKAQAYVRSLPFRRTVPFRRVLPGSDPLALDLMAKMLSFDPSTR VTVLESLEHPWLAAYHDESDEPECSERFERWREIEKLETLDDFRKALWDEIEDYRREVRGLLPVVDPLELKERDA DKERDATKEEEFSETKAPEGDVQNGKDTPSADVSVSTVPSEPAVETPPTEPTAPVAVVDTSLKSTAASIISANKV MVDSPESQHVQTSKGGTDPVVTYARRSSIMQPSRQGSTYNSPVPSTQYLPTFIEGQSLNLGGDAASILGASGIGA GAGNSIAFPSTQPPGYVVPARSRTGSTVGGEVNRRLLRTLSTVSIHEPSQGKVGGLAGVAPIGKFIVRGDGEGVT EGVTEADAPPSEMPREFLGRSGEGSGSGDETKTKGDKKERKKFHVE |
| Length | 721 |
Coding
| Sequence id | CopciAB_379729.T0 |
|---|---|
| Sequence |
>CopciAB_379729.T0 ATGTCTCGAGTAAGAGGTGGCCATTCTCCTGCACGATCCCCGTCTCGACCTCATCGGCGGAGGTCAACCCGTGGT CGGTCCAACGGTGACGAGAACGGCAATGGCGATGGCAGCAACGACAACAACAAATCGTCTTCACCTGTACCCTCT CCCGGCTCTTGGAATGTGGATCGAGGACGATCGAGGGTGAGGAGGCCGTCCAACCCCTTCTCGGAGGATAATCTG CGTCGAAGAGGGTACCATTCGCTGCATTCATCGTTTGGCAAAGTGTTTCATGTAGAGAAGAGGTGGAAGCTGGTG AGGGAGATGGGTTCTGGTGCTTACGGCGTGGTAATTTCAGCGGCCGACGAAATCTCAGGCGAAACAGTAGCCATC AAGATGGTCACCCGAGTGTTTGAAAAGATACAACTTGCAAAACGGGCTCTCAGGGAGATCACCCTCCTCAGACAT TTCGCCAACCACGAAAATATAACTGGCTTGATTGACGTAGATGCTGTTTCCCCTGATTTCAATGAAATTTACATC TTCATGGAGCCCATGGAAGCCGACCTTCACCAAATCATCAAGTCTGGTCAAGTTCTCACCAACGAACATGTGCAA TATTTCTTGTACCAGATTCTGCGAGGAATGAAATTCATTCACTCTGCGGGAGTCATCCATCGAGACCTCAAACCG GGCAATCTGCTCGTGAACGCTGATTGCGAACTCAAGATATGCGATTTTGGCCTTAGCAGGGGGTTCGATGCTGTC CCCGACGAGAACGCTACGCGACTTACGGAGTACGTGGCGACGAGGTGGTACAGGGCGCCGGAGATCATGTTGGCA TTTCCACGATATAATACTGCTATCGATGTTTGGTCAATTGGCTGCATTCTGGGAGAACTACTGCTAGGAAAACCC CTCTTTAAGGGGAAAGATTACGTTGACCAACTAAACAAAATCCTGGACATACTAGGCTCGCCGGACGAGACTATC ATCCAAAAGATTGGGAGCGACAAGGCTCAAGCCTATGTCCGTTCTTTACCGTTTAGGAGAACTGTTCCATTCCGA CGCGTTCTACCTGGCTCGGACCCGCTAGCCCTTGACTTGATGGCCAAAATGCTCTCGTTTGATCCATCTACGCGT GTAACGGTTCTCGAATCCCTCGAACACCCATGGCTCGCTGCATATCACGACGAAAGCGACGAACCTGAATGTTCT GAACGGTTTGAAAGGTGGAGAGAGATCGAAAAACTGGAGACATTGGACGACTTCCGCAAGGCGCTGTGGGATGAA ATCGAGGACTACCGTCGGGAAGTCCGGGGGCTCTTGCCTGTGGTTGACCCGCTGGAACTGAAGGAACGGGACGCG GACAAGGAGAGGGACGCGACCAAGGAAGAGGAGTTTTCAGAGACGAAGGCTCCAGAAGGCGATGTTCAGAATGGG AAAGATACTCCGAGTGCAGATGTTTCGGTTTCCACAGTACCTTCAGAACCTGCCGTCGAGACACCGCCCACCGAG CCGACTGCGCCCGTGGCTGTCGTGGACACGAGCTTGAAGTCGACTGCAGCTTCCATCATCAGCGCCAACAAAGTC ATGGTCGACAGCCCAGAATCACAACACGTCCAAACCTCGAAAGGCGGAACGGACCCAGTCGTGACGTATGCTCGT CGTTCGAGCATCATGCAACCCTCGAGACAAGGATCGACGTACAACTCTCCCGTTCCGTCGACGCAGTATCTCCCG ACCTTCATCGAAGGGCAGAGCCTGAACCTCGGTGGAGACGCGGCCAGCATACTCGGGGCTTCAGGGATCGGTGCA GGGGCCGGAAACTCCATTGCTTTCCCAAGTACCCAGCCACCTGGCTACGTCGTCCCTGCACGATCGCGAACCGGA TCCACTGTTGGTGGGGAGGTTAATAGGAGGTTGCTCCGCACCCTTTCGACGGTGTCTATCCATGAACCAAGTCAG GGCAAAGTGGGTGGTTTGGCAGGTGTGGCTCCGATTGGAAAGTTCATTGTTAGGGGAGACGGTGAGGGCGTCACG GAGGGCGTCACGGAGGCTGATGCACCTCCAAGTGAAATGCCGAGAGAGTTCTTGGGTAGGAGCGGGGAGGGCAGT GGGAGTGGAGATGAAACCAAGACAAAGGGAGACAAAAAGGAAAGGAAAAAATTCCATGTCGAATAA |
| Length | 2166 |
Transcript
| Sequence id | CopciAB_379729.T0 |
|---|---|
| Sequence |
>CopciAB_379729.T0 ACCTTTTTGTTTCTTCTCGCGACTGTGTTCAGCCTTCTACCACATTTCCCATCTACTTGGGGGGACGAGTCTATT TCAGTCAGCTACCTGGTCTGAATTCCTTTGCCACGCGGTCCAGTGCTGTGTGGTCCTTGGGTCGCTGGGAAGCTC AAACGTTCCAGGGCTGTGCGTGTCGAACGTTGACGACCGCGATACACACCTTGGACGAGAATGTCTCGAGTAAGA GGTGGCCATTCTCCTGCACGATCCCCGTCTCGACCTCATCGGCGGAGGTCAACCCGTGGTCGGTCCAACGGTGAC GAGAACGGCAATGGCGATGGCAGCAACGACAACAACAAATCGTCTTCACCTGTACCCTCTCCCGGCTCTTGGAAT GTGGATCGAGGACGATCGAGGGTGAGGAGGCCGTCCAACCCCTTCTCGGAGGATAATCTGCGTCGAAGAGGGTAC CATTCGCTGCATTCATCGTTTGGCAAAGTGTTTCATGTAGAGAAGAGGTGGAAGCTGGTGAGGGAGATGGGTTCT GGTGCTTACGGCGTGGTAATTTCAGCGGCCGACGAAATCTCAGGCGAAACAGTAGCCATCAAGATGGTCACCCGA GTGTTTGAAAAGATACAACTTGCAAAACGGGCTCTCAGGGAGATCACCCTCCTCAGACATTTCGCCAACCACGAA AATATAACTGGCTTGATTGACGTAGATGCTGTTTCCCCTGATTTCAATGAAATTTACATCTTCATGGAGCCCATG GAAGCCGACCTTCACCAAATCATCAAGTCTGGTCAAGTTCTCACCAACGAACATGTGCAATATTTCTTGTACCAG ATTCTGCGAGGAATGAAATTCATTCACTCTGCGGGAGTCATCCATCGAGACCTCAAACCGGGCAATCTGCTCGTG AACGCTGATTGCGAACTCAAGATATGCGATTTTGGCCTTAGCAGGGGGTTCGATGCTGTCCCCGACGAGAACGCT ACGCGACTTACGGAGTACGTGGCGACGAGGTGGTACAGGGCGCCGGAGATCATGTTGGCATTTCCACGATATAAT ACTGCTATCGATGTTTGGTCAATTGGCTGCATTCTGGGAGAACTACTGCTAGGAAAACCCCTCTTTAAGGGGAAA GATTACGTTGACCAACTAAACAAAATCCTGGACATACTAGGCTCGCCGGACGAGACTATCATCCAAAAGATTGGG AGCGACAAGGCTCAAGCCTATGTCCGTTCTTTACCGTTTAGGAGAACTGTTCCATTCCGACGCGTTCTACCTGGC TCGGACCCGCTAGCCCTTGACTTGATGGCCAAAATGCTCTCGTTTGATCCATCTACGCGTGTAACGGTTCTCGAA TCCCTCGAACACCCATGGCTCGCTGCATATCACGACGAAAGCGACGAACCTGAATGTTCTGAACGGTTTGAAAGG TGGAGAGAGATCGAAAAACTGGAGACATTGGACGACTTCCGCAAGGCGCTGTGGGATGAAATCGAGGACTACCGT CGGGAAGTCCGGGGGCTCTTGCCTGTGGTTGACCCGCTGGAACTGAAGGAACGGGACGCGGACAAGGAGAGGGAC GCGACCAAGGAAGAGGAGTTTTCAGAGACGAAGGCTCCAGAAGGCGATGTTCAGAATGGGAAAGATACTCCGAGT GCAGATGTTTCGGTTTCCACAGTACCTTCAGAACCTGCCGTCGAGACACCGCCCACCGAGCCGACTGCGCCCGTG GCTGTCGTGGACACGAGCTTGAAGTCGACTGCAGCTTCCATCATCAGCGCCAACAAAGTCATGGTCGACAGCCCA GAATCACAACACGTCCAAACCTCGAAAGGCGGAACGGACCCAGTCGTGACGTATGCTCGTCGTTCGAGCATCATG CAACCCTCGAGACAAGGATCGACGTACAACTCTCCCGTTCCGTCGACGCAGTATCTCCCGACCTTCATCGAAGGG CAGAGCCTGAACCTCGGTGGAGACGCGGCCAGCATACTCGGGGCTTCAGGGATCGGTGCAGGGGCCGGAAACTCC ATTGCTTTCCCAAGTACCCAGCCACCTGGCTACGTCGTCCCTGCACGATCGCGAACCGGATCCACTGTTGGTGGG GAGGTTAATAGGAGGTTGCTCCGCACCCTTTCGACGGTGTCTATCCATGAACCAAGTCAGGGCAAAGTGGGTGGT TTGGCAGGTGTGGCTCCGATTGGAAAGTTCATTGTTAGGGGAGACGGTGAGGGCGTCACGGAGGGCGTCACGGAG GCTGATGCACCTCCAAGTGAAATGCCGAGAGAGTTCTTGGGTAGGAGCGGGGAGGGCAGTGGGAGTGGAGATGAA ACCAAGACAAAGGGAGACAAAAAGGAAAGGAAAAAATTCCATGTCGAATAAGCGGTGGAGGGAGGAATAGTGCCA CGCGCCGTGACGGAGGCGATTCTCAGCGTAATTTCTCTTAACTGATCTCTACAAACGTACCTAGTTTATCAGTTT AATGTCGTAATATTTCTTACATCCGTAGTTTGATTTTATTATAGGTTGTTGAAATGGGAGAACGCAGAGTTGGAC CGTCAC |
| Length | 2556 |
Gene
| Sequence id | CopciAB_379729.T0 |
|---|---|
| Sequence |
>CopciAB_379729.T0 ACCTTTTTGTTTCTTCTCGCGACTGTGTTCAGCCTTCTACCACATTTCCCATCTACTTGGGGGGACGAGTCTATT TCAGTCAGCTACCTGGTCTGAATTCCTTTGCCACGCGGTCCAGTGCTGTGTGGTCCTTGGGTCGCTGGGAAGCTC AAACGTTCCAGGGCTGTGCGTGTCGAACGTTGACGACCGCGATACACACCTTGGACGAGAATGTCTCGAGTAAGA GGTGGCCATTCTCCTGCACGATCCCCGTCTCGACCTCATCGGCGGAGGTCAACCCGTGGTCGGTCCAACGGTGAC GAGAACGGCAATGGCGATGGCAGCAACGACAACAACAAATCGTCTTCACCTGTACCCTCTCCCGGCTCTTGGAAT GTGGATCGAGGACGATCGAGGGTGAGGAGGCCGTCCAACCCCTTCTCGGAGGATAATCTGCGTCGAAGGTGCGTA GGTGTTGGCACACGACACCTTTGCTGTCAATGTTCACCTTGTTCCTGACTGTATTCCTTGGATGTAATGAAGAGG GTACCATTCGCTGCATTCATCGTTTGGCAAAGTGTTTCATGTAGAGAAGAGGTGGAAGCTGGTGAGGGAGATGGG TTCTGGTGCTTACGGCGTGGTAATGTAGGTCAGTCTTGCTTTGCCTCTGAGGCTCGTTATCGTCTAATGCAGGTG TTATTTCTCCTTCCCTGGGTCCATACGAAACCTGCCCCCATCCCCTCGCCCTCCAGTTCAGCGGCCGACGAAATC TCAGGCGAAACAGTAGCCATCAAGATGGTCACCCGAGTGTTTGAAAAGATACAACTTGCAAAACGGGCTCTCAGG GAGATCACCCTCCTCAGACATTTCGCCAACCACGAAAATATAACTGGCTTGATTGACGTAGATGCTGTTTCCCCT GATTTCAATGAAATGTGAGTTGGTTTATACCTGGTTCGCCATTATCAAAGGGACTGACGTCATTATTTCCGTTCA ATAGTTACATCTTCATGGAGGTGCGTCGGACGGTATTGCAAGGTTGGGAAGATTCTAAAGGCGGTGAACATTGGC GGTTTAGCCCATGGAAGGTGTTTGCAACGTGCTGTTGAATGAGATGACCCTATTGAAACGCGTTCATACTTCCTA GCCGACCTTCACCAAATCATCAAGTCTGGTCAAGTTCTCACCAACGAACATGTGCAATATTTCTTGTACCAGATT CTGCGAGGTACCATGATAACTCGTCTGTTATATCTCGCTGTTGCTGATCCTTCAACTTTAAGGAATGAAATTCAT TCACTCTGCGGGAGTCATCCATCGAGACCTCAAACCGGGCAATCTGCTCGTGAACGCTGATTGCGAACTCAAGAT ATGCGATTTTGGCCTTAGCAGGGGGTTCGATGCTGTCCCCGACGAGAACGCTACGCGACTTACGGAGTACGTGGC GACGAGGTGGTACAGGGCGCCGGAGATCATGTTGGCATTTCCGTGAGTGTTCTACGCTTGGCCCGAGATTGGGTA TCTGACTGCGGTTGTCAGACGATATAATACTGCTAGTAAATCATATCAGCTTGCCTTGAATTTCTTGTGCTTACT GGGTATCCTAGTCGATGTTTGGTCAATTGGCTGCATTCTGGGAGAACTACTGCTAGGAAAACCCCTCTTTAAGGG GAAAGAGTGAGTACACACCCTGTTTGATGTGTCACGCTCTGATTGCTTTTCAGTTACGGTTCGCCATTTTCTGTA AAACCTTTGCGACTCCTCTGACCATGCTACTGCAGTTGACCAACTAAACAAAATCCTGGACATACTAGGCTCGCC GGACGAGACTATCATCCAAAAGATTGGGAGCGACAAGGTCAGGGAGATCTAACTCTCACCGTTGACTGTTGGTTC ACCGATATGGTTAGGCTCAAGCCTATGTCCGTTCTTTACCGTTTAGGAGAACTGTTCCATTCCGACGCGTTCTAC CTGGCTCGGACCCGCTAGGTACAAGCGATTTCCTTCAGGCCGGAACTAATGATCGCTGACTTGCATCGTAGCCCT TGACTTGATGGCCAAAATGCTCTCGTTTGATCCATCTACGCGTGTAACGGTTCTCGAATCCCTCGAACACCCATG GCTCGCTGCATATCACGACGAAAGCGACGAACCTGAATGTTCTGAACGGTTTGAAAGGTGGAGAGAGATCGAAAA ACTGGAGACATTGGACGACTTCCGCAAGGCGCTGTGGGATGAAATCGAGGACTACCGTCGGGAAGTCCGGGGGCT CTTGCCTGTGGTTGACCCGCTGGAACTGAAGGAACGGGACGCGGACAAGGAGAGGGACGCGACCAAGGAAGAGGA GTTTTCAGAGACGAAGGCTCCAGAAGGCGATGTTCAGAATGGGAAAGATACTCCGAGTGCAGATGTTTCGGTTTC CACAGTACCTTCAGAACCTGCCGTCGAGACACCGCCCACCGAGCCGACTGCGCCCGTGGCTGTCGTGGACACGAG CTTGAAGTCGACTGCAGCTTCCATCATCAGCGCCAACAAAGTCATGGTCGACAGCCCAGAATCACAACACGTCCA AACCTCGAAAGGCGGAACGGACCCAGTCGTGACGTATGCTCGTCGTTCGAGCATCATGCAACCCTCGAGACAAGG ATCGACGTACAACTCTCCCGTTCCGTCGACGCAGTATCTCCCGACCTTCATCGAAGGGCAGAGCCTGAACCTCGG TGGAGACGCGGCCAGCATACTCGGGGCTTCAGGGATCGGTGCAGGGGCCGGAAACTCCATTGCTTTCCCAAGTAC CCAGCCACCTGGCTACGTCGTCCCTGCACGATCGCGAACCGGATCCACTGTTGGTGGGGAGGTTAATAGGAGGTT GCTCCGCACCCTTTCGACGGTGTCTATCCATGAACCAAGTCAGGGCAAAGTGGGTGGTTTGGCAGGTGTGGCTCC GATTGGAAAGTTCATTGTTAGGGGAGACGGTGAGGGCGTCACGGAGGGCGTCACGGAGGCTGATGCACCTCCAAG TGAAATGCCGAGAGAGTTCTTGGGTAGGAGCGGGGAGGGCAGTGGGAGTGGAGATGAAACCAAGACAAAGGGAGA CAAAAAGGAAAGGAAAAAATTCCATGTCGAATAAGCGGTGGAGGGAGGAATAGTGCCACGCGCCGTGACGGAGGC GATTCTCAGCGTAATTTCTCTTAACTGATCTCTACAAACGTACCTAGTTTATCAGTTTAATGTCGTAATATTTCT TACATCCGTAGTTTGATTTTATTATAGGTTGTTGAAATGGGAGAACGCAGAGTTGGACCGTCAC |
| Length | 3289 |