CopciAB_393662
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_393662 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 393662 |
| Uniprot id | Functional description | Other 1 protein kinase | |
| Location | scaffold_5:137781..140687 | Strand | + |
| Gene length (nt) | 2907 | Transcript length (nt) | 2620 |
| CDS length (nt) | 2448 | Protein length (aa) | 815 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Flammulina velutipes | Flave_chr08AA00131 | 29.8 | 6.775E-72 | 261 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1443306 | 25.5 | 4.973E-41 | 164 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14846 | 25.6 | 2.487E-36 | 149 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87994 | 26.3 | 4.268E-33 | 138 |
| Pleurotus ostreatus PC15 | PleosPC15_2_170780 | 26.1 | 1.721E-28 | 123 |
| Agrocybe aegerita | Agrae_CAA7264594 | 24.2 | 1.184E-25 | 114 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_23187 | 25.8 | 8.584E-21 | 98 |
| Lentinula edodes NBRC 111202 | Lenedo1_1165864 | 25.4 | 5.83E-19 | 92 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_393662.T0 |
| Description | Other 1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 585 | 694 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR040976 | Fungal-type protein kinase |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| E | Other 1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
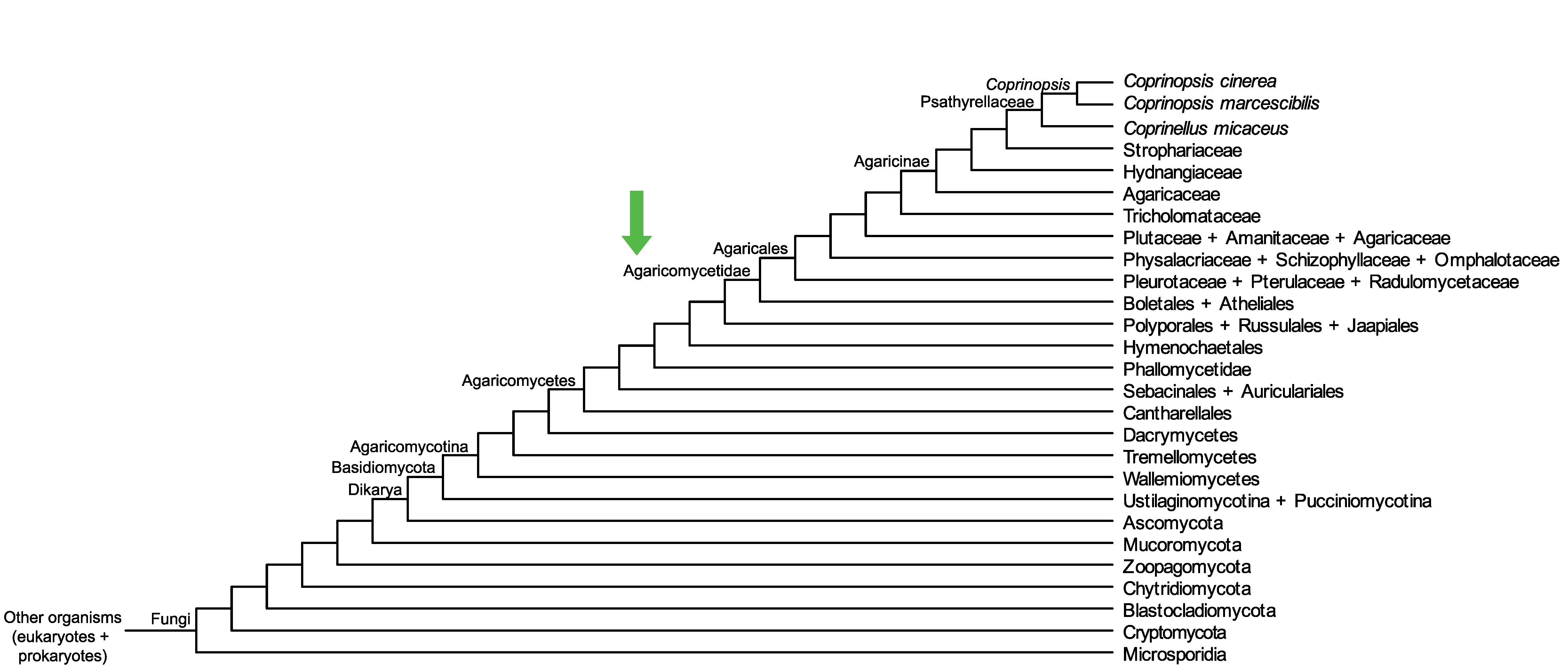
Conservation of CopciAB_393662 across fungi.
Arrow shows the origin of gene family containing CopciAB_393662.
Protein
| Sequence id | CopciAB_393662.T0 |
|---|---|
| Sequence |
>CopciAB_393662.T0 METNSEYTTEGLGPWCALRRSRLFTSVAGSEITCGCIGFQAPSIIQLVEDDLEDTLHTQLKDMLTAFLRKMRIDG PNFDAQNEFNAFLDESVRFCNSNSAEAKAVKASILEAMRCSTEKDRYPHLAYALNSILKHGRGKTIGSFNHSLDK DESLMAVNDPAIVQCGDTVLSASECTDDYYLKRNPALLRMPIQALGRLVGRNEPFDVLLTRLATSKSRLEEKNAN WKDIIDCWEIRKGTDLIPEHSFKRLTDEEYKFTVECADERPFDKASVPIPATTNTSSTRPSAPPPSSTGTSSPSQ TSVNLSSGILKRPRTAEDSQSDNAHTAHKRPKTEPESDPEPNPLHLRLSPETQVGVYALELMRTRWNRTHAIVIL LEGNKLSLHWYDPQGCIRTHPIDIVSQLPLLVAMMLIFQRFDDRMRGNGKFQLNIDIDGETVPHSLDDSEIHNRW MFKGGRSVSGKLFPHGKIPPPKSESRSSRTTRNNANPTANDVPSTEEGELFGKFLWREEARHTEGVIIETALKRA ERHLGEHKKHVIDHLPCVKTCTEFSDFSTKHIRNFLNLPSQGARVPTGIAAKWLLPSDELTDPGEYVRAIWQLIR CNFLLWQLGIAHGDISMTNLMLRPGSGSVQAVLNDFDLAAIMTPGNTSPERKGFERTGTIPFMAVELLRATNGEV KRLWCHDFESFLWCMVWYCSPAVRSIWDSGTAPQVGNEKLGWLASEPYTSRSKDTTVAEAAKPIWTSLRKLFRGW DYSFFTFGQENTGWEEQLAFLHRLVEHMSEYDPCEDRSWLEFRGKPSMQCVCSPPSSAVPCSTCT |
| Length | 815 |
Coding
| Sequence id | CopciAB_393662.T0 |
|---|---|
| Sequence |
>CopciAB_393662.T0 ATGGAGACCAATTCAGAATATACCACGGAAGGTTTAGGGCCTTGGTGCGCACTGCGTCGTAGTCGACTATTCACC TCGGTGGCAGGATCTGAAATCACTTGTGGATGTATAGGTTTTCAAGCCCCGTCCATCATCCAACTTGTTGAGGAC GATCTGGAGGATACTCTGCATACCCAGCTCAAGGATATGCTTACTGCGTTTCTGAGGAAGATGCGGATAGACGGG CCCAATTTCGACGCCCAGAACGAGTTCAACGCATTCCTGGACGAATCCGTCAGGTTTTGTAACAGCAACAGTGCC GAGGCGAAAGCCGTGAAAGCCAGCATTCTCGAAGCAATGCGATGTTCGACCGAAAAGGATCGCTATCCCCACCTG GCATACGCTCTCAACAGCATCTTGAAGCATGGCCGAGGGAAGACTATTGGAAGTTTCAACCACAGCCTGGATAAA GACGAATCCCTTATGGCCGTGAATGATCCCGCCATCGTGCAATGCGGCGACACCGTCCTATCTGCCAGTGAATGC ACCGATGATTATTACTTGAAGCGAAACCCCGCCCTGCTTCGCATGCCGATACAAGCGCTGGGACGCCTTGTGGGC AGGAACGAACCTTTCGACGTCTTGTTGACGCGTCTTGCCACCTCCAAGAGCAGATTGGAGGAAAAGAACGCAAAT TGGAAGGACATCATCGACTGTTGGGAGATCAGGAAGGGCACAGACCTCATTCCCGAGCACAGCTTTAAGCGCCTG ACAGACGAGGAATACAAATTCACAGTTGAATGTGCCGATGAACGACCTTTCGACAAGGCGAGCGTCCCCATTCCC GCGACTACTAATACCTCGAGCACACGTCCCTCGGCCCCACCGCCTTCTTCCACTGGGACAAGTTCTCCTAGCCAA ACTTCAGTCAACCTATCCTCTGGAATCCTCAAACGACCGCGGACAGCGGAGGACTCACAGTCCGATAACGCCCAC ACCGCTCACAAGAGACCCAAAACAGAACCAGAATCTGACCCAGAACCAAACCCTCTCCACCTCCGTCTAAGCCCG GAAACCCAGGTTGGCGTCTACGCGCTCGAGCTCATGCGAACAAGGTGGAACCGCACGCACGCTATCGTCATCCTT CTGGAAGGCAATAAACTATCCCTCCACTGGTACGACCCACAGGGATGCATCAGAACCCACCCTATCGACATCGTC AGCCAGCTTCCTCTCCTTGTCGCCATGATGCTCATCTTCCAGCGCTTCGACGATCGGATGCGTGGCAACGGCAAG TTCCAGTTGAACATCGATATCGACGGGGAGACAGTCCCACACAGCCTCGACGACTCGGAGATCCACAATCGCTGG ATGTTCAAAGGCGGTCGTTCAGTCAGTGGGAAGCTGTTCCCGCACGGAAAGATACCACCACCGAAGTCAGAATCA CGCAGCTCTCGAACGACCCGCAACAATGCCAACCCCACGGCCAACGATGTACCCTCGACTGAGGAGGGGGAACTG TTTGGCAAATTCTTGTGGCGTGAAGAGGCCCGGCACACTGAAGGAGTCATCATTGAGACTGCCCTGAAGAGGGCC GAACGACACCTCGGCGAACACAAGAAACATGTCATCGACCACCTCCCTTGCGTTAAAACCTGCACCGAATTTTCC GACTTTTCGACCAAGCACATCCGAAATTTCCTGAACCTACCTTCCCAAGGTGCTCGGGTGCCAACTGGAATAGCT GCTAAATGGTTGCTTCCAAGTGACGAACTCACAGACCCGGGGGAGTACGTTCGTGCTATCTGGCAGCTCATTCGA TGCAATTTCCTACTGTGGCAACTCGGGATCGCGCACGGCGATATCAGCATGACGAACCTCATGCTTCGCCCGGGC AGTGGTAGCGTTCAGGCTGTTCTAAATGACTTCGACTTGGCGGCGATCATGACCCCAGGAAATACTTCCCCCGAG AGGAAAGGTTTCGAGCGGACGGGAACCATACCCTTCATGGCGGTTGAGCTCTTGCGGGCCACGAATGGAGAAGTC AAGCGCCTTTGGTGCCACGATTTCGAATCCTTCCTCTGGTGCATGGTGTGGTACTGCTCGCCGGCGGTGCGGAGC ATCTGGGACTCAGGCACCGCTCCCCAAGTGGGCAACGAGAAGTTGGGCTGGCTCGCCTCCGAGCCTTACACAAGT AGAAGTAAAGACACCACCGTTGCCGAAGCAGCCAAGCCTATTTGGACGTCGCTGAGGAAACTCTTTCGCGGTTGG GACTACAGCTTCTTCACTTTTGGTCAGGAGAACACTGGGTGGGAGGAGCAACTCGCGTTCCTGCATAGATTGGTC GAGCACATGTCAGAGTACGATCCATGCGAGGATCGGAGCTGGTTGGAGTTTCGGGGAAAGCCATCCATGCAGTGC GTGTGTAGTCCTCCTTCCTCTGCTGTTCCCTGTTCGACCTGTACTTGA |
| Length | 2448 |
Transcript
| Sequence id | CopciAB_393662.T0 |
|---|---|
| Sequence |
>CopciAB_393662.T0 AGAGAGACGCCGTCAGGTCCGAAACTGGGTCTACCCCCCTAAACAGGGAATATGGAGACCAATTCAGAATATACC ACGGAAGGTTTAGGGCCTTGGTGCGCACTGCGTCGTAGTCGACTATTCACCTCGGTGGCAGGATCTGAAATCACT TGTGGATGTATAGGTTTTCAAGCCCCGTCCATCATCCAACTTGTTGAGGACGATCTGGAGGATACTCTGCATACC CAGCTCAAGGATATGCTTACTGCGTTTCTGAGGAAGATGCGGATAGACGGGCCCAATTTCGACGCCCAGAACGAG TTCAACGCATTCCTGGACGAATCCGTCAGGTTTTGTAACAGCAACAGTGCCGAGGCGAAAGCCGTGAAAGCCAGC ATTCTCGAAGCAATGCGATGTTCGACCGAAAAGGATCGCTATCCCCACCTGGCATACGCTCTCAACAGCATCTTG AAGCATGGCCGAGGGAAGACTATTGGAAGTTTCAACCACAGCCTGGATAAAGACGAATCCCTTATGGCCGTGAAT GATCCCGCCATCGTGCAATGCGGCGACACCGTCCTATCTGCCAGTGAATGCACCGATGATTATTACTTGAAGCGA AACCCCGCCCTGCTTCGCATGCCGATACAAGCGCTGGGACGCCTTGTGGGCAGGAACGAACCTTTCGACGTCTTG TTGACGCGTCTTGCCACCTCCAAGAGCAGATTGGAGGAAAAGAACGCAAATTGGAAGGACATCATCGACTGTTGG GAGATCAGGAAGGGCACAGACCTCATTCCCGAGCACAGCTTTAAGCGCCTGACAGACGAGGAATACAAATTCACA GTTGAATGTGCCGATGAACGACCTTTCGACAAGGCGAGCGTCCCCATTCCCGCGACTACTAATACCTCGAGCACA CGTCCCTCGGCCCCACCGCCTTCTTCCACTGGGACAAGTTCTCCTAGCCAAACTTCAGTCAACCTATCCTCTGGA ATCCTCAAACGACCGCGGACAGCGGAGGACTCACAGTCCGATAACGCCCACACCGCTCACAAGAGACCCAAAACA GAACCAGAATCTGACCCAGAACCAAACCCTCTCCACCTCCGTCTAAGCCCGGAAACCCAGGTTGGCGTCTACGCG CTCGAGCTCATGCGAACAAGGTGGAACCGCACGCACGCTATCGTCATCCTTCTGGAAGGCAATAAACTATCCCTC CACTGGTACGACCCACAGGGATGCATCAGAACCCACCCTATCGACATCGTCAGCCAGCTTCCTCTCCTTGTCGCC ATGATGCTCATCTTCCAGCGCTTCGACGATCGGATGCGTGGCAACGGCAAGTTCCAGTTGAACATCGATATCGAC GGGGAGACAGTCCCACACAGCCTCGACGACTCGGAGATCCACAATCGCTGGATGTTCAAAGGCGGTCGTTCAGTC AGTGGGAAGCTGTTCCCGCACGGAAAGATACCACCACCGAAGTCAGAATCACGCAGCTCTCGAACGACCCGCAAC AATGCCAACCCCACGGCCAACGATGTACCCTCGACTGAGGAGGGGGAACTGTTTGGCAAATTCTTGTGGCGTGAA GAGGCCCGGCACACTGAAGGAGTCATCATTGAGACTGCCCTGAAGAGGGCCGAACGACACCTCGGCGAACACAAG AAACATGTCATCGACCACCTCCCTTGCGTTAAAACCTGCACCGAATTTTCCGACTTTTCGACCAAGCACATCCGA AATTTCCTGAACCTACCTTCCCAAGGTGCTCGGGTGCCAACTGGAATAGCTGCTAAATGGTTGCTTCCAAGTGAC GAACTCACAGACCCGGGGGAGTACGTTCGTGCTATCTGGCAGCTCATTCGATGCAATTTCCTACTGTGGCAACTC GGGATCGCGCACGGCGATATCAGCATGACGAACCTCATGCTTCGCCCGGGCAGTGGTAGCGTTCAGGCTGTTCTA AATGACTTCGACTTGGCGGCGATCATGACCCCAGGAAATACTTCCCCCGAGAGGAAAGGTTTCGAGCGGACGGGA ACCATACCCTTCATGGCGGTTGAGCTCTTGCGGGCCACGAATGGAGAAGTCAAGCGCCTTTGGTGCCACGATTTC GAATCCTTCCTCTGGTGCATGGTGTGGTACTGCTCGCCGGCGGTGCGGAGCATCTGGGACTCAGGCACCGCTCCC CAAGTGGGCAACGAGAAGTTGGGCTGGCTCGCCTCCGAGCCTTACACAAGTAGAAGTAAAGACACCACCGTTGCC GAAGCAGCCAAGCCTATTTGGACGTCGCTGAGGAAACTCTTTCGCGGTTGGGACTACAGCTTCTTCACTTTTGGT CAGGAGAACACTGGGTGGGAGGAGCAACTCGCGTTCCTGCATAGATTGGTCGAGCACATGTCAGAGTACGATCCA TGCGAGGATCGGAGCTGGTTGGAGTTTCGGGGAAAGCCATCCATGCAGTGCGTGTGTAGTCCTCCTTCCTCTGCT GTTCCCTGTTCGACCTGTACTTGATTTGGCTCCGGTCAAATTCCTTGTGTTCGATGGAAACGTTGAGAGTCCGTA GCGTTAGGTCCTGTTCACTATTCTTCCTGTGATTGAGATTTGTTACAAATATAATCTATGATTCCGGTTC |
| Length | 2620 |
Gene
| Sequence id | CopciAB_393662.T0 |
|---|---|
| Sequence |
>CopciAB_393662.T0 AGAGAGACGCCGTCAGGTCCGAAACTGGGTCTACCCCCCTAAACAGGGAATATGGAGACCAATTCAGAATATACC ACGGAAGGTTTAGGGCCTTGGTGCGCACTGCGTCGTAGTCGACTATTCACCTCGGTGGCAGGATCTGAAATCACT TGTGGATGTATAGGTTTTCAAGCCCCGTCCATCATCCAACTTGTTGAGGACGATCTGGAGGATACTCTGCATACC CAGCTCAAGGATATGCTTACTGCGTTTCTGAGGAAGATGCGGATAGACGGGCCCAATTTCGACGCCCAGAACGAG TTCAACGCATTCCTGGACGAATCCGTCAGGTTTTGTAACAGCAACAGTGCCGAGGCGAAAGCCGTGAAAGCCAGC ATTCTCGAAGCGTATGTCCATCAGTACTTGTATACCCACCTACGATTACTAACTCTCTGCTTTGTCTTTCAGAAT GCGATGTTCGACCGAAAAGGATCGCTATCCCCACCTGGCATACGCTCTCAACAGCATCTTGAAGCATGGCCGAGG GAAGACTATTGGAAGTTTCAACCACAGCCTGGATAAAGACGAATCCCTTATGGCCGTGAATGATCCCGCCATCGT GCAATGCGGCGACACCGTCCTATCTGCCAGTGAATGCACCGATGATTATTACTTGAAGCGAAACCCCGCCCTGCT TCGCATGCCGATACAAGCGCTGGGACGCCTTGTGGGCAGGAACGAACCTTTCGACGTCTTGTTGACGCGTCTTGC CACCTCCAAGAGCAGATTGGAGGAAAAGAACGCAAATTGGAAGGACATCATCGACTGTTGGGAGATCAGGAAGGG CACAGACCTCATTCCCGAGCACAGCTTTAAGCGCCTGACAGACGAGGAATACAAATTCACAGTTGAATGTGCCGA TGAACGACGTGAGTTGACAACCTTTCTCTCTCTCTCTCTCTCTCTCGGATTGGTTTTCAGCTAAATTCGAATCAC TTTTGTTTGTATGTAGCTTTCGACAAGGCGAGCGTCCCCATTCCCGCGACTACTAATACCTCGAGCACACGTCCC TCGGCCCCACCGCCTTCTTCCACTGGGACAAGTTCTCCTAGCCAAACTTCAGTCAACCTATCCTCTGGAATCCTC AAACGACCGCGGACAGCGGAGGACTCACAGTCCGATAACGCCCACACCGCTCACAAGAGACCCAAAACAGAACCA GAATCTGACCCAGAACCAAACCCTCTCCACCTCCGTCTAAGCCCGGAAACCCAGGTTGGCGTCTACGCGCTCGAG CTCATGCGAACAAGGTGGAACCGCACGCACGCTATCGTCATCCTTCTGGAAGGTAGGCGCATCATATCCCTTCCA ATTGCAACAGCTCCTAATTATTTGATTCGTCATTTTATAGGCAATAAACTATCCCTCCACTGGTACGACCCACAG GGATGCATCAGAACCCACCCTATCGACATCGTCAGCCAGCTTCCTCTCCTTGTCGCCATGATGCTCATCTTCCAG CGCTTCGACGATCGGATGCGTGGCAACGGCAAGTTCCAGTTGAACATCGATATCGACGGGGAGACAGTCCCACAC AGCCTCGACGACTCGGAGATCCACAATCGCTGGATGTTCAAAGGCGGTCGTTCAGTCAGTGGGAAGCTGTTCCCG CACGGAAAGATACCACCACCGAAGTCAGAATCACGCAGCTCTCGAACGACCCGCAACAATGCCAACCCCACGGCC AACGATGTACCCTCGACTGAGGAGGGGGAACTGTTTGGCAAATTCTTGTGGCGTGAAGAGGCCCGGCACACTGAA GGAGTCATCATTGAGACTGCCCTGAAGAGGGCCGAACGACACCTCGGCGAACACAAGAAACATGTCATCGACCAC CTCCCTTGCGTTAAAACCTGCACCGAATTTTCCGACTTTTCGACCAAGCACATCCGAAATTTCCTGAACCTACCT TCCCAAGGTGCTCGGGTGCCAACTGGAATAGCTGCTAAATGGTTGCTTCCAAGTGACGAACTCACAGACCCGGGG GAGTACGTTCGTGCTATCTGGCAGCTCATTCGATGTAAGTACTACTTCATGCGCGTTGTACTAGTACCTCCCCAT TCAATCGAGCTAACGTCCACTTCCTCTGCTCTCTCCTAGGCAATTTCCTACTGTGGCAACTCGGGATCGCGCACG GCGATATCAGCATGACGAACCTCATGCTTCGCCCGGGCAGTGGTAGCGTTCAGGCTGTTCTAAATGACTTCGACT TGGCGGCGATCATGACCCCAGGAAATACTTCCCCCGAGAGGAAAGGTTTCGAGCGGACGGGAACCATACCCTTCA TGGCGGTTGAGCTCTTGCGGGCCACGAATGGAGAAGTCAAGCGCCTTTGGTGCCACGATTTCGAATCCTTCCTCT GGTGCATGGTGTGGTACTGCTCGCCGGCGGTGCGGAGCATCTGGGACTCAGGCACCGCTCCCCAAGTGGGCAACG AGAAGTTGGGCTGGCTCGCCTCCGAGCCTTACACAAGTAGAAGTAAAGACACCACCGTTGCCGAAGCAGCCAAGC CTATTTGGACGTCGCTGAGGAAACTCTTTCGCGGTTGGGACTACAGCTTCTTCACTTTTGGTCAGGAGAACACTG GGTGGGAGGAGCAACTCGCGTTCCTGCATAGATTGGTCGAGCACATGTCAGAGTACGATCCATGCGAGGATCGGA GCTGGTTGGAGTTTCGGGGAAAGCCATCCATGCAGTGCGTGTGTAGTCCTCCTTCCTCTGCTGTTCCCTGTTCGA CCTGTACTTGATTTGGCTCCGGTCAAATTCCTTGTGTTCGATGGAAACGTTGAGAGTCCGTAGCGTTAGGTCCTG TTCACTATTCTTCCTGTGATTGAGATTTGTTACAAATATAATCTATGATTCCGGTTC |
| Length | 2907 |