CopciAB_403152
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_403152 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | CDC5 | Synonyms | 403152 |
| Uniprot id | Functional description | Kinase-like | |
| Location | scaffold_8:2137619..2141151 | Strand | + |
| Gene length (nt) | 3533 | Transcript length (nt) | 3223 |
| CDS length (nt) | 2859 | Protein length (aa) | 952 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | plkA |
| Neurospora crassa | cdc5 |
| Candida albicans | cdc5 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7261230 | 66.3 | 0 | 1212 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB27078 | 65.4 | 0 | 1155 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_76212 | 62.8 | 0 | 1071 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_188194 | 59.8 | 0 | 1060 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_42993 | 59.8 | 0 | 1059 |
| Grifola frondosa | Grifr_OBZ65532 | 58.4 | 0 | 1037 |
| Pleurotus ostreatus PC15 | PleosPC15_2_37936 | 57.3 | 0 | 1020 |
| Pleurotus ostreatus PC9 | PleosPC9_1_88693 | 57.3 | 0 | 1020 |
| Schizophyllum commune H4-8 | Schco3_17460 | 58.8 | 0 | 1014 |
| Lentinula edodes NBRC 111202 | Lenedo1_1167217 | 60.1 | 0 | 1013 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_9190 | 60.1 | 0 | 1013 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1392086 | 56.1 | 0 | 1011 |
| Flammulina velutipes | Flave_chr10AA01084 | 58.2 | 4.614E-304 | 987 |
| Auricularia subglabra | Aurde3_1_1326196 | 55.2 | 3.107E-278 | 875 |
| Lentinula edodes B17 | Lened_B_1_1_9615 | 81.5 | 6.308E-69 | 254 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | CDC5 |
|---|---|
| Protein id | CopciAB_403152.T0 |
| Description | Kinase-like |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14099 | STKc_PLK | - | 80 | 336 |
| CDD | cd13118 | POLO_box_1 | IPR033701 | 656 | 743 |
| CDD | cd13117 | POLO_box_2 | IPR033695 | 762 | 851 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 83 | 336 |
| Pfam | PF00659 | POLO box duplicated region | IPR000959 | 667 | 729 |
| Pfam | PF00659 | POLO box duplicated region | IPR000959 | 769 | 822 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000959 | POLO box domain |
| IPR000719 | Protein kinase domain |
| IPR017441 | Protein kinase, ATP binding site |
| IPR036947 | POLO box domain superfamily |
| IPR033701 | First polo-box domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR033695 | Second polo-box domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005515 | protein binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K06631 |
| K06660 |
EggNOG
| COG category | Description |
|---|---|
| D | Kinase-like |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
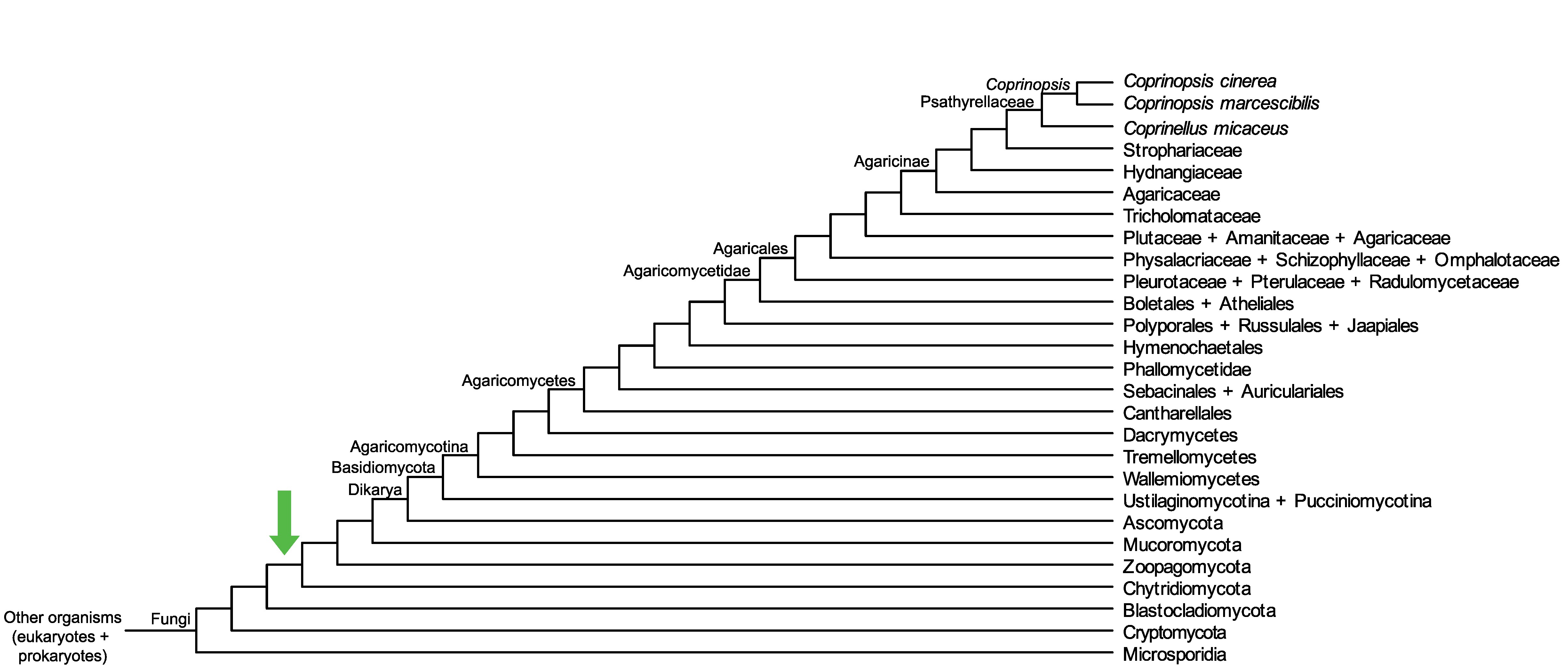
Conservation of CopciAB_403152 across fungi.
Arrow shows the origin of gene family containing CopciAB_403152.
Protein
| Sequence id | CopciAB_403152.T0 |
|---|---|
| Sequence |
>CopciAB_403152.T0 MYQMNGYRMPPAAPGPSSKRNPLVNAVNHHPNVMQMNAVPPVPAKAPAKGPSSPPLPRQNSKTRPPSPPKVISDK AGRLQFNRVGFLGEGGFARVYEVQDPRGSRLACKVVTKSALKTKKAKTKLYAEIKIHRSLDHPNIVTFIDCFEDE DNVYITLELCPSGSLMDMLRRRRRFTEPETRFFMVQLIGACHYMHTHQVIHRDLKLGNLFLDANMNVKVGDFGLA ALIENPGERKKTICGTPNYIAPEVLFDTANGHSFEVDIWSIGVILYTLVIGRPPFQTKEVKEIYKRIRDNEYEFP AERAISTAVKVLIQQILTPDPSQRPTLHEIVEDTFFTHGTVPAFIPTSAHDAAPDFRHISRSASEANLKRLRRYA LLDTDFNNTPSPVQNPTTRNITSSIAQQEKEFQRAVQPGSPISALLSSARQPLLVGPTAGGGGNPGAVRESPLLR KLQAVKESPLGRRSVTRTANGYEEDERGKRRMVIETEEEEEDMRQRRKELEAQKARIVAQMAPVKEEVEREERED EDDEEEDEDGLGGRRRAPPPPPPVFKREREHQKGSSAGSNRYAGEDVENVPPRLGGAQQARRDVAKGKAVERPRA REKENVPVSSSNPAPAAPKLNGFDAAAQILTMAFDARAAGRVFRSPLGDQPLPEERVFIVSWVDYCNKYGMGYAL TDGSVGVHFNDSTSLVLSADKNHFDYISSRHHGSIYLRKSFTIDAYPDELKSKVYLLKHFEKYIMERLYGEYEYT FEDVERTKGMEWVQKYLRMKHVIVFKMSHDVLQFNFYDHSKLILSSNGLLITHIDKSYKMTRWTLEDVMAFSLSP PAGASGEEIKFNQRMVDKLKYCKEVLVSIRSATSAGAGGGGERGTSTERGEKERGEKERGGERAERGERRMGGDD ERERGGGERERERERERERADRGVERERLGSGREAGSSTATLNRRTSKLSMR |
| Length | 952 |
Coding
| Sequence id | CopciAB_403152.T0 |
|---|---|
| Sequence |
>CopciAB_403152.T0 ATGTATCAGATGAATGGGTATCGAATGCCACCCGCCGCTCCTGGCCCAAGCTCGAAACGAAACCCGCTGGTCAAC GCCGTCAACCACCACCCGAATGTTATGCAGATGAACGCCGTACCGCCTGTGCCAGCCAAAGCCCCCGCCAAGGGT CCGTCGTCCCCGCCGCTGCCGAGACAGAACAGCAAGACGAGACCGCCGTCGCCACCCAAGGTCATCAGCGACAAG GCTGGTCGACTGCAGTTTAACCGTGTTGGATTCCTGGGAGAGGGAGGGTTTGCTCGGGTGTATGAAGTCCAGGAT CCGCGAGGAAGTCGACTTGCTTGCAAGGTAGTTACAAAGTCTGCTCTCAAGACCAAGAAAGCTAAAACCAAGCTC TACGCAGAAATCAAGATCCACCGCTCACTCGACCACCCCAACATCGTCACGTTCATCGACTGTTTTGAAGACGAA GACAATGTCTACATCACCCTCGAGCTGTGTCCTTCTGGATCACTCATGGACATGCTGCGTCGTCGCAGACGCTTC ACAGAACCAGAAACAAGGTTTTTCATGGTACAGCTCATAGGAGCGTGCCACTACATGCACACCCACCAGGTCATC CACAGAGACCTCAAACTCGGAAACCTCTTCCTCGACGCCAACATGAACGTCAAGGTCGGCGACTTTGGTCTCGCT GCCCTCATCGAGAACCCCGGTGAGCGAAAGAAGACCATCTGCGGAACACCCAACTACATCGCACCCGAGGTCTTG TTCGACACCGCCAACGGTCACAGCTTTGAGGTCGACATTTGGTCGATCGGTGTCATTCTCTACACCCTCGTCATC GGACGCCCGCCTTTCCAGACAAAGGAGGTCAAGGAGATCTACAAACGTATCCGTGACAACGAGTACGAGTTCCCC GCTGAGCGGGCCATCTCCACCGCCGTCAAGGTTCTCATTCAGCAAATCCTCACCCCAGATCCATCTCAGCGCCCC ACCCTCCACGAGATCGTCGAAGACACTTTCTTCACCCACGGCACCGTCCCCGCCTTCATTCCCACCTCTGCCCAC GACGCCGCACCCGACTTCCGCCACATCTCCCGCAGCGCCTCCGAAGCCAACCTCAAACGCCTCCGCCGCTACGCC CTCCTCGACACCGACTTTAATAACACGCCCTCCCCTGTTCAGAACCCCACGACGAGGAACATTACGTCGAGTATC GCCCAGCAGGAGAAGGAATTCCAGCGGGCTGTTCAGCCTGGTTCGCCTATTTCTGCGTTGCTCAGTTCGGCGAGG CAGCCGTTGTTGGTGGGTCCTACAGCGGGTGGTGGCGGGAACCCCGGAGCTGTACGTGAGAGCCCGCTTTTGCGG AAGCTGCAGGCTGTCAAGGAGAGTCCGTTGGGGAGGCGGAGTGTCACTCGCACTGCCAACGGGTACGAGGAGGAT GAGAGGGGTAAGAGGCGGATGGTGATCGAGACGGAGGAGGAAGAGGAGGATATGAGGCAGAGGAGGAAGGAGCTT GAAGCGCAGAAGGCGAGGATTGTGGCGCAGATGGCGCCTGTTAAGGAGGAGGTGGAGAGGGAGGAGAGGGAGGAT GAAGACGACGAGGAGGAAGATGAGGATGGGTTGGGTGGGAGGAGGAGGGCGCCGCCGCCTCCGCCGCCTGTGTTT AAGAGGGAGAGGGAGCATCAGAAGGGGTCGTCGGCGGGTTCGAATCGGTATGCGGGGGAGGATGTCGAGAATGTG CCGCCGAGGTTGGGTGGGGCGCAGCAGGCGAGGAGGGATGTGGCGAAGGGGAAGGCTGTTGAGAGGCCGAGGGCG AGGGAGAAGGAGAATGTTCCCGTGTCGTCTTCGAATCCAGCACCTGCAGCACCCAAACTGAACGGCTTTGACGCG GCAGCTCAGATCCTCACTATGGCGTTTGACGCGCGGGCTGCGGGCCGCGTGTTCCGTAGTCCGTTGGGCGATCAG CCGCTGCCTGAGGAGCGGGTGTTTATCGTGAGCTGGGTGGATTATTGCAACAAGTATGGGATGGGGTATGCACTT ACCGACGGGAGTGTTGGTGTGCATTTCAATGATAGTACGAGTTTGGTGCTGAGTGCTGACAAGAACCATTTCGAC TACATCTCCTCCCGCCACCATGGATCCATCTACCTTCGCAAATCCTTCACGATCGACGCCTATCCCGACGAGTTG AAGAGTAAAGTCTACCTTTTGAAGCATTTTGAAAAGTATATTATGGAGCGGTTGTATGGGGAGTATGAGTATACG TTTGAGGATGTTGAGAGGACGAAGGGGATGGAGTGGGTGCAAAAGTATTTGAGGATGAAGCATGTTATTGTGTTT AAGATGAGTCATGATGTTTTGCAGTTCAACTTTTACGACCACTCGAAGCTCATCCTCTCCTCCAACGGTCTTCTC ATCACGCACATTGACAAATCGTACAAGATGACGCGGTGGACGCTTGAAGATGTTATGGCGTTTTCACTATCGCCA CCCGCTGGGGCGAGTGGGGAGGAGATCAAGTTTAATCAGAGGATGGTGGATAAGTTGAAGTATTGTAAAGAGGTT TTGGTTAGTATTAGGAGTGCTACTAGTGCTGGGGCTGGTGGGGGAGGGGAGCGGGGAACGTCGACGGAGCGGGGG GAGAAGGAGAGGGGGGAGAAGGAGAGGGGAGGGGAGAGGGCTGAACGGGGGGAGAGGAGGATGGGTGGGGATGAT GAGAGGGAGAGGGGTGGTGGGGAGAGGGAGAGGGAGAGGGAGAGGGAGAGGGAGAGGGCGGATAGAGGCGTGGAG AGGGAGAGGCTTGGCTCTGGGAGGGAAGCTGGTTCGTCTACTGCCACTCTGAATCGGAGGACGTCCAAGTTGTCT ATGCGGTAG |
| Length | 2859 |
Transcript
| Sequence id | CopciAB_403152.T0 |
|---|---|
| Sequence |
>CopciAB_403152.T0 ATCCTCGTTGTTTGTTTTCGCTCCATCGCTTGCTCGCTCGCCGTCCGCAAACCGCCAGTCAGTGGTGGCCTTTGC GATTTTTTCCCTTTCTTTCTCAATTCTAGGGCGTTAGGCCTTTCAAGCTCAAGATTTTAACACCCCGCCAAAAAA ATCCGAATATGTATCAGATGAATGGGTATCGAATGCCACCCGCCGCTCCTGGCCCAAGCTCGAAACGAAACCCGC TGGTCAACGCCGTCAACCACCACCCGAATGTTATGCAGATGAACGCCGTACCGCCTGTGCCAGCCAAAGCCCCCG CCAAGGGTCCGTCGTCCCCGCCGCTGCCGAGACAGAACAGCAAGACGAGACCGCCGTCGCCACCCAAGGTCATCA GCGACAAGGCTGGTCGACTGCAGTTTAACCGTGTTGGATTCCTGGGAGAGGGAGGGTTTGCTCGGGTGTATGAAG TCCAGGATCCGCGAGGAAGTCGACTTGCTTGCAAGGTAGTTACAAAGTCTGCTCTCAAGACCAAGAAAGCTAAAA CCAAGCTCTACGCAGAAATCAAGATCCACCGCTCACTCGACCACCCCAACATCGTCACGTTCATCGACTGTTTTG AAGACGAAGACAATGTCTACATCACCCTCGAGCTGTGTCCTTCTGGATCACTCATGGACATGCTGCGTCGTCGCA GACGCTTCACAGAACCAGAAACAAGGTTTTTCATGGTACAGCTCATAGGAGCGTGCCACTACATGCACACCCACC AGGTCATCCACAGAGACCTCAAACTCGGAAACCTCTTCCTCGACGCCAACATGAACGTCAAGGTCGGCGACTTTG GTCTCGCTGCCCTCATCGAGAACCCCGGTGAGCGAAAGAAGACCATCTGCGGAACACCCAACTACATCGCACCCG AGGTCTTGTTCGACACCGCCAACGGTCACAGCTTTGAGGTCGACATTTGGTCGATCGGTGTCATTCTCTACACCC TCGTCATCGGACGCCCGCCTTTCCAGACAAAGGAGGTCAAGGAGATCTACAAACGTATCCGTGACAACGAGTACG AGTTCCCCGCTGAGCGGGCCATCTCCACCGCCGTCAAGGTTCTCATTCAGCAAATCCTCACCCCAGATCCATCTC AGCGCCCCACCCTCCACGAGATCGTCGAAGACACTTTCTTCACCCACGGCACCGTCCCCGCCTTCATTCCCACCT CTGCCCACGACGCCGCACCCGACTTCCGCCACATCTCCCGCAGCGCCTCCGAAGCCAACCTCAAACGCCTCCGCC GCTACGCCCTCCTCGACACCGACTTTAATAACACGCCCTCCCCTGTTCAGAACCCCACGACGAGGAACATTACGT CGAGTATCGCCCAGCAGGAGAAGGAATTCCAGCGGGCTGTTCAGCCTGGTTCGCCTATTTCTGCGTTGCTCAGTT CGGCGAGGCAGCCGTTGTTGGTGGGTCCTACAGCGGGTGGTGGCGGGAACCCCGGAGCTGTACGTGAGAGCCCGC TTTTGCGGAAGCTGCAGGCTGTCAAGGAGAGTCCGTTGGGGAGGCGGAGTGTCACTCGCACTGCCAACGGGTACG AGGAGGATGAGAGGGGTAAGAGGCGGATGGTGATCGAGACGGAGGAGGAAGAGGAGGATATGAGGCAGAGGAGGA AGGAGCTTGAAGCGCAGAAGGCGAGGATTGTGGCGCAGATGGCGCCTGTTAAGGAGGAGGTGGAGAGGGAGGAGA GGGAGGATGAAGACGACGAGGAGGAAGATGAGGATGGGTTGGGTGGGAGGAGGAGGGCGCCGCCGCCTCCGCCGC CTGTGTTTAAGAGGGAGAGGGAGCATCAGAAGGGGTCGTCGGCGGGTTCGAATCGGTATGCGGGGGAGGATGTCG AGAATGTGCCGCCGAGGTTGGGTGGGGCGCAGCAGGCGAGGAGGGATGTGGCGAAGGGGAAGGCTGTTGAGAGGC CGAGGGCGAGGGAGAAGGAGAATGTTCCCGTGTCGTCTTCGAATCCAGCACCTGCAGCACCCAAACTGAACGGCT TTGACGCGGCAGCTCAGATCCTCACTATGGCGTTTGACGCGCGGGCTGCGGGCCGCGTGTTCCGTAGTCCGTTGG GCGATCAGCCGCTGCCTGAGGAGCGGGTGTTTATCGTGAGCTGGGTGGATTATTGCAACAAGTATGGGATGGGGT ATGCACTTACCGACGGGAGTGTTGGTGTGCATTTCAATGATAGTACGAGTTTGGTGCTGAGTGCTGACAAGAACC ATTTCGACTACATCTCCTCCCGCCACCATGGATCCATCTACCTTCGCAAATCCTTCACGATCGACGCCTATCCCG ACGAGTTGAAGAGTAAAGTCTACCTTTTGAAGCATTTTGAAAAGTATATTATGGAGCGGTTGTATGGGGAGTATG AGTATACGTTTGAGGATGTTGAGAGGACGAAGGGGATGGAGTGGGTGCAAAAGTATTTGAGGATGAAGCATGTTA TTGTGTTTAAGATGAGTCATGATGTTTTGCAGTTCAACTTTTACGACCACTCGAAGCTCATCCTCTCCTCCAACG GTCTTCTCATCACGCACATTGACAAATCGTACAAGATGACGCGGTGGACGCTTGAAGATGTTATGGCGTTTTCAC TATCGCCACCCGCTGGGGCGAGTGGGGAGGAGATCAAGTTTAATCAGAGGATGGTGGATAAGTTGAAGTATTGTA AAGAGGTTTTGGTTAGTATTAGGAGTGCTACTAGTGCTGGGGCTGGTGGGGGAGGGGAGCGGGGAACGTCGACGG AGCGGGGGGAGAAGGAGAGGGGGGAGAAGGAGAGGGGAGGGGAGAGGGCTGAACGGGGGGAGAGGAGGATGGGTG GGGATGATGAGAGGGAGAGGGGTGGTGGGGAGAGGGAGAGGGAGAGGGAGAGGGAGAGGGAGAGGGCGGATAGAG GCGTGGAGAGGGAGAGGCTTGGCTCTGGGAGGGAAGCTGGTTCGTCTACTGCCACTCTGAATCGGAGGACGTCCA AGTTGTCTATGCGGTAGTTGCCAATGGTGGTGTGTGTGGTGTTTTTTATGTGTGTAACCCCCTCGCGTTCTCTGC TACCCTTCTTTGTCACCCCTCGCGTTCTCTATCTCTACCCTTCTTTGTCGCTGTCTCGTTTGTTAAGTACTACTC TACTCTTTTGTGTGTCGCTATCGCGTTTTTGTGTTCGTTTGGTATTCTAGACTCGATTTTTATTATGCGCTCC |
| Length | 3223 |
Gene
| Sequence id | CopciAB_403152.T0 |
|---|---|
| Sequence |
>CopciAB_403152.T0 ATCCTCGTTGTTTGTTTTCGCTCCATCGCTTGCTCGCTCGCCGTCCGCAAACCGCCAGTCAGTGGTGGCCTTTGC GATTTTTTCCCTTTCTTTCTCAATTCTAGGGCGTTAGGCCTTTCAAGCTCAAGATTTTAACACCCCGCCAAAAAA ATCCGAATATGTATCAGATGAATGGGTATCGAATGCCACCCGCCGCTCCTGGCCCAAGCTCGAAACGAAACCCGC TGGTCAACGCCGTCAACCACCACCCGAATGTTATGCAGATGAACGCCGTACCGCCTGTGCCAGCCAAAGCCCCCG CCAAGGGTCCGTCGTCCCCGCCGCTGCCGAGACAGAACAGCAAGACGAGACCGCCGTCGCCACCCAAGGTCATCA GCGACAAGGCTGGTCGACTGCAGTTTAACCGTGTTGGATTCCTGGGAGAGGTGAGAGGTTAGCTAAGTATATTTG GGTTGGACTAACGACTGTGTAGGGAGGGTTTGCTCGGGTGTATGAAGTCCAGGATCCGCGAGGAAGTCGACTTGC TTGCAAGGTAGTTACAAAGTCTGCTCTCAAGACCAAGAAAGCTAAAACCAAGGTGAGTGTTTTCTGATACTCTCC AGTCGTTTCTAATCCCTTTTAGCTCTACGCAGAAATCAAGATCCACCGCTCACTCGACCACCCCAACATCGTCAC GTTCATCGACTGTTTTGAAGACGAAGACAATGTCTACATCACCCTCGAGCTGTGTCCTTCTGGATCACTCATGGA CATGCTGCGTCGTCGCAGACGCTTCACAGAACCAGAAACAAGGTTTTTCATGGTACAGCTCATAGGAGCGTGCCA CTACATGCACACCCACCAGGTCATCCACAGAGACCTCAAACTCGGAAACCTCTTCCTCGACGCCAACATGAACGT CAAGGTCGGCGACTTTGGTCTCGCTGCCCTCATCGAGAACCCCGGTGAGCGAAAGAAGACCATCTGCGGAACACC CAACTACATCGCACCCGAGGTCTTGTTCGACACCGCCAACGGTCACAGCTTTGAGGTCGACATTTGGTCGATCGG TGTCATTCTCTACACCCTCGTCATCGGACGCCCGCCTTTCCAGACAAAGGAGGTCAAGGAGATCTACAAGTAAGT CTTGCTTTTATCGTCTTTCGCCTTGTTCTAATAATGGTTCTAGACGTATCCGTGACAACGAGTACGAGTTCCCCG CTGAGCGGGCCATCTCCACCGCCGTCAAGGTTCTCATTCAGCAAATCCTCACCCCAGATCCATCTCAGCGCCCCA CCCTCCACGAGATCGTCGAAGACACTTTCTTCACCCACGGCACCGTCCCCGCCTTCATTCCCACCTCTGCCCACG ACGCCGCACCCGACTTCCGCCACATCTCCCGCAGCGCCTCCGAAGCCAACCTCAAACGCCTCCGCCGCTACGCCC TCCTCGACACCGACTTTAATAACACGCCCTCCCCTGTTCAGAACCCCACGACGAGGAACATTACGTCGAGTATCG CCCAGCAGGAGAAGGAATTCCAGCGGGCTGTTCAGCCTGGTTCGCCTATTTCTGCGTTGCTCAGTTCGGCGAGGC AGCCGTTGTTGGTGGGTCCTACAGCGGGTGGTGGCGGGAACCCCGGAGCTGTACGTGAGAGCCCGCTTTTGCGGA AGCTGCAGGCTGTCAAGGAGAGTCCGTTGGGGAGGCGGAGTGTCACTCGCACTGCCAACGGGTACGAGGAGGATG AGAGGGGTAAGAGGCGGATGGTGATCGAGACGGAGGAGGAAGAGGAGGATATGAGGCAGAGGAGGAAGGAGCTTG AAGCGCAGAAGGCGAGGATTGTGGCGCAGATGGCGCCTGTTAAGGAGGAGGTGGAGAGGGAGGAGAGGGAGGATG AAGACGACGAGGAGGAAGATGAGGATGGGTTGGGTGGGAGGAGGAGGGCGCCGCCGCCTCCGCCGCCTGTGTTTA AGAGGGAGAGGGAGCATCAGAAGGGGTCGTCGGCGGGTTCGAATCGGTATGCGGGGGAGGATGTCGAGAATGTGC CGCCGAGGTTGGGTGGGGCGCAGCAGGCGAGGAGGGATGTGGCGAAGGGGAAGGCTGTTGAGAGGCCGAGGGCGA GGGAGAAGGAGAATGTTCCCGTGTCGTGTATGTCCCATTTCTGTCCATGATACCATGAAGCTAACTCATCTCTAG CTTCGAATCCAGCACCTGCAGCACCCAAACTGAACGGCTTTGACGCGGCAGCTCAGATCCTCACTATGGCGTTTG ACGCGCGGGCTGCGGGCCGCGTGTTCCGTAGTCCGTTGGGCGATCAGCCGCTGCCTGAGGAGCGGGTGTTTATCG TGAGCTGGGTGGATTATTGCAACAAGTATGGGATGGGGTATGCACTTACCGACGGGAGTGTTGGTGTGCATTTCA ATGATAGTACGAGTTTGGTGCTGAGTGCTGACAAGAAGTGGGTCTCTCCTCTCTCTCGTGTCGTGTTGTCGCTGA CTAATGTTCGCCAACAGCCATTTCGACTACATCTCCTCCCGCCACCATGGATCCATCTACCTTCGCAAATCCTTC ACGATCGACGCCTATCCCGACGAGTTGAAGAGTAAAGTCTACCTTTTGAAGCATTTTGAAAAGTATATTATGGAG CGGTTGTATGGGGAGTATGAGTATACGTTTGAGGATGTTGAGAGGACGAAGGGGATGGAGTGGGTGCAAAAGTAT TTGAGGATGAAGCATGTTATTGTGTTTAAGATGAGTCATGATGTTTTGCAGGTGCGTCCCCCTCTCTCTCTCGCC GTCTCGTTACCTTACTGACGCACCACCTTTCGTCGACTCTAGTTCAACTTTTACGACCACTCGAAGCTCATCCTC TCCTCCAACGGTCTTCTCATCACGCACATTGACAAATCGTACAAGATGACGCGGTGGACGCTTGAAGATGTTATG GCGTTTTCACTATCGCCACCCGCTGGGGCGAGTGGGGAGGAGATCAAGTTTAATCAGAGGATGGTGGATAAGTTG AAGTATTGTAAAGAGGTTTTGGTTAGTATTAGGAGTGCTACTAGTGCTGGGGCTGGTGGGGGAGGGGAGCGGGGA ACGTCGACGGAGCGGGGGGAGAAGGAGAGGGGGGAGAAGGAGAGGGGAGGGGAGAGGGCTGAACGGGGGGAGAGG AGGATGGGTGGGGATGATGAGAGGGAGAGGGGTGGTGGGGAGAGGGAGAGGGAGAGGGAGAGGGAGAGGGAGAGG GCGGATAGAGGCGTGGAGAGGGAGAGGCTTGGCTCTGGGAGGGAAGCTGGTTCGTCTACTGCCACTCTGAATCGG AGGACGTCCAAGTTGTCTATGCGGTAGTTGCCAATGGTGGTGTGTGTGGTGTTTTTTATGTGTGTAACCCCCTCG CGTTCTCTGCTACCCTTCTTTGTCACCCCTCGCGTTCTCTATCTCTACCCTTCTTTGTCGCTGTCTCGTTTGTTA AGTACTACTCTACTCTTTTGTGTGTCGCTATCGCGTTTTTGTGTTCGTTTGGTATTCTAGACTCGATTTTTATTA TGCGCTCC |
| Length | 3533 |