CopciAB_404912
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_404912 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 404912 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_12:242805..245733 | Strand | + |
| Gene length (nt) | 2929 | Transcript length (nt) | 2338 |
| CDS length (nt) | 2211 | Protein length (aa) | 736 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB18405 | 43.5 | 1.083E-92 | 322 |
| Agrocybe aegerita | Agrae_CAA7268614 | 43.2 | 4.555E-91 | 317 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_78633 | 40.3 | 1.585E-80 | 285 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_208652 | 40.1 | 1.507E-79 | 282 |
| Pleurotus eryngii ATCC 90797 | Pleery1_274204 | 33.5 | 2.008E-58 | 218 |
| Schizophyllum commune H4-8 | Schco3_2016992 | 35.7 | 2.262E-58 | 218 |
| Lentinula edodes B17 | Lened_B_1_1_9770 | 28.7 | 6.493E-57 | 213 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1045430 | 36.2 | 3.102E-56 | 211 |
| Pleurotus ostreatus PC9 | PleosPC9_1_57702 | 36.2 | 3.036E-56 | 211 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_1618 | 35.7 | 3.319E-55 | 208 |
| Lentinula edodes NBRC 111202 | Lenedo1_84152 | 28.3 | 5.901E-53 | 201 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_404912.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00071 | Ras family | IPR001806 | 13 | 167 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 435 | 700 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR005225 | Small GTP-binding protein domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR001806 | Small GTPase |
| IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005525 | GTP binding | MF |
| GO:0003924 | GTPase activity | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
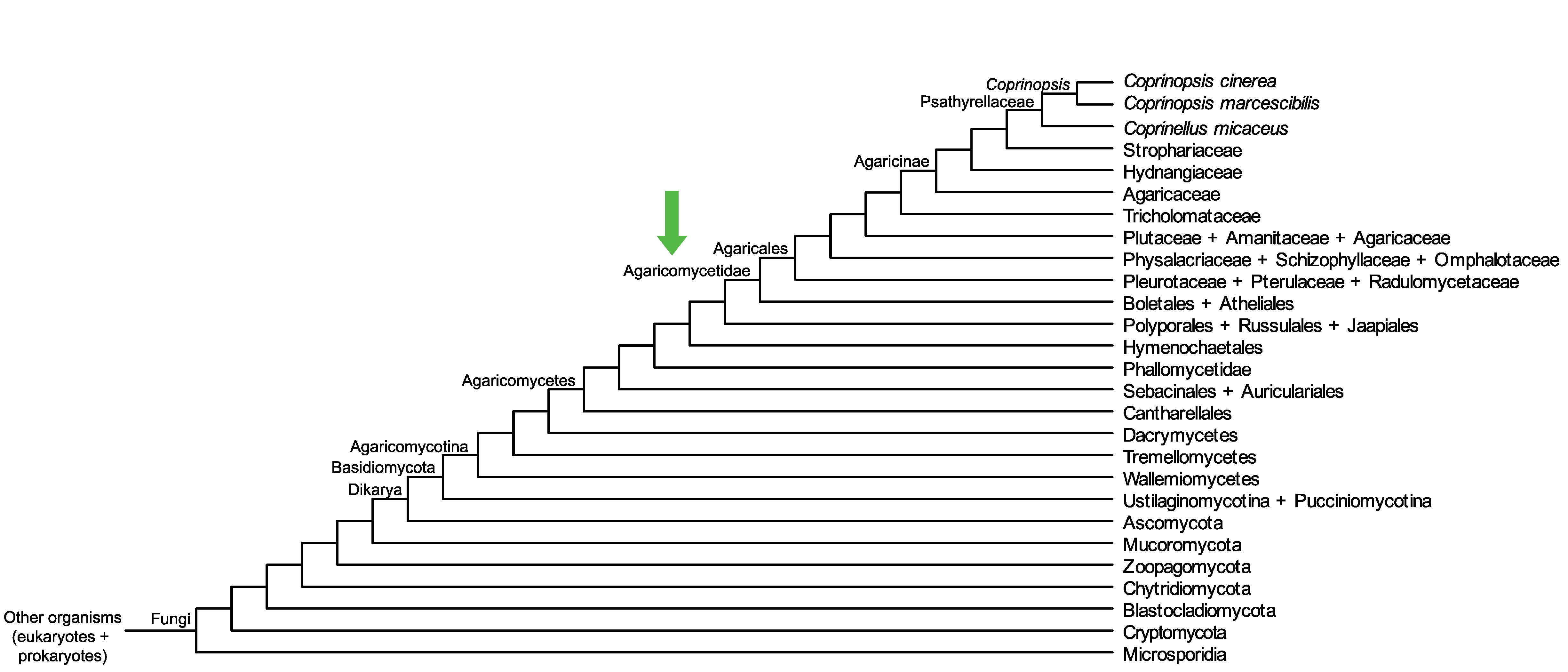
Conservation of CopciAB_404912 across fungi.
Arrow shows the origin of gene family containing CopciAB_404912.
Protein
| Sequence id | CopciAB_404912.T0 |
|---|---|
| Sequence |
>CopciAB_404912.T0 MREFQTTKKRLNIVFLGGRRVGKTSLLSQWSQGEFPSAYYPTFENTVGKEVTFGNRIYDCKVTDTAGHDRYTPLS PTHVVGMHVFVLVYSFEDAGSLELVSALYEKILEFLGSTRHARVIMAGNKADTSRREISYPEAHQLAQKLGCPHL ILSSYWWQSIDQLFEECTIGGERRNRQPPTRTSATAKPQSRGLWFPSIDLLSSLLRYASRVTWPSSAANTTSIPE PTDTYHQIQRHTKPHSEEELHGSQAPIPITTPLDYDRPTRSSVRGYSIMSTSSGEATLVSTFGPKSRLSASSSKG VSPFQKRRSFSPMTPIKLNVTKDDVVKSKFRFVDELTRIQRVRGVDAGLAQDFTNLFRSIMEDKVTLKTLLEFRG HDAQVVLNALQWVLDSRKAQPADKGMLLKLLLRLSKHSGSYPPCLALKGIKREDDYPVTSGHFGEVWRGRYQKQK VCLKVVRVYQRSDVEHFLRIFSREAVIWSQLKHDNVLPFYGVYELDESRRLCLVSPWMDNGSLREYVRKSLLSEV HHRLLAVDVTQGLSYLHQQGMVHGDLKGDNILIDSSGRACIADFGLSSLVDGEMLRWTTSESTGLGVGGTSRWQA PELFDPDNESSRPTCYTDVYSLGCVFYEIFTGNIPFHETTREATVISYVMAGKKPSRPVNNYILYRMEYGMREII WSTIEKCWSRDVSTRPSLDRILSLEVFQDAVRKDLRPNGHLSWLSTSEFRHAIRFTENDFL |
| Length | 736 |
Coding
| Sequence id | CopciAB_404912.T0 |
|---|---|
| Sequence |
>CopciAB_404912.T0 ATGAGAGAGTTTCAGACAACGAAGAAGCGTCTCAATATTGTCTTCCTTGGTGGCCGTCGGGTTGGGAAGACATCG TTGTTGTCGCAATGGTCTCAGGGAGAATTCCCGAGTGCATATTATCCGACGTTCGAGAACACTGTGGGGAAGGAG GTGACATTTGGGAACCGAATCTACGACTGTAAGGTCACTGATACAGCGGGCCATGATCGATATACCCCGCTGTCT CCTACCCATGTTGTTGGTATGCACGTATTTGTGCTGGTTTACTCGTTTGAAGACGCCGGATCGTTGGAATTGGTC TCGGCTCTCTACGAGAAGATCTTGGAATTTTTAGGGTCCACCAGACATGCCCGAGTTATCATGGCTGGGAACAAG GCCGATACATCTCGACGAGAGATATCCTATCCAGAGGCCCACCAACTCGCACAGAAACTGGGCTGCCCTCATCTA ATATTGAGTAGCTACTGGTGGCAAAGTATCGATCAACTATTCGAAGAGTGTACTATAGGCGGTGAACGGAGAAAC CGCCAGCCTCCAACCCGCACATCAGCAACAGCCAAGCCTCAATCTCGTGGCTTGTGGTTCCCAAGCATCGACTTA CTATCTTCCCTGCTTCGCTACGCTTCTCGCGTCACCTGGCCCAGCTCCGCGGCAAACACCACCTCAATACCAGAG CCCACTGACACCTACCATCAGATTCAACGTCACACCAAGCCACACAGCGAGGAGGAATTGCATGGCTCACAGGCC CCCATCCCAATTACGACGCCATTGGATTACGATCGTCCAACGCGTAGTTCAGTCAGGGGCTACTCAATAATGTCA ACCTCATCCGGCGAAGCCACCCTCGTATCTACGTTTGGTCCAAAGTCACGCCTATCGGCCTCATCCAGCAAGGGA GTTAGTCCATTCCAGAAACGGCGCTCTTTTAGCCCCATGACTCCGATCAAGCTCAATGTCACCAAGGACGATGTC GTCAAGAGCAAATTTCGGTTCGTTGATGAACTAACGAGGATACAGAGGGTGCGGGGTGTTGACGCGGGACTTGCA CAAGACTTTACGAACCTATTTCGCTCGATTATGGAAGATAAAGTGACGCTGAAGACCTTGCTGGAATTTCGGGGA CATGATGCTCAGGTGGTTCTGAACGCTCTGCAATGGGTTCTGGATTCGCGGAAAGCTCAGCCGGCGGATAAAGGA ATGTTGCTGAAGCTGCTTCTTCGACTGTCGAAGCATTCTGGGTCCTATCCTCCGTGTCTGGCGCTGAAAGGAATT AAGCGCGAAGACGACTATCCGGTTACATCAGGACACTTTGGTGAGGTTTGGAGGGGTCGATACCAGAAGCAGAAA GTTTGCTTGAAGGTGGTCAGGGTGTATCAGCGATCGGATGTTGAGCACTTCCTTCGGATATTCTCTCGCGAAGCT GTGATCTGGAGTCAACTGAAGCACGACAATGTTCTTCCTTTCTATGGCGTCTATGAACTCGACGAGTCTCGTCGA CTTTGCCTGGTCTCTCCTTGGATGGATAACGGAAGTCTTCGGGAGTATGTGCGAAAGAGCTTGTTGTCGGAGGTC CACCATCGCCTTCTGGCAGTCGATGTAACACAGGGCCTGTCCTATCTTCATCAGCAGGGAATGGTCCATGGCGAT CTTAAAGGGGATAACATCCTCATTGACAGTTCTGGACGTGCCTGCATAGCGGACTTCGGGCTCTCAAGTCTGGTG GACGGCGAGATGCTTCGTTGGACAACATCCGAGAGTACCGGACTAGGCGTTGGTGGCACATCCCGCTGGCAAGCT CCAGAACTGTTCGATCCCGATAATGAATCGAGTCGGCCCACGTGTTATACGGATGTTTATTCGCTTGGTTGCGTT TTCTACGAGATCTTCACTGGAAACATCCCATTCCACGAGACAACACGCGAAGCCACAGTGATATCGTACGTCATG GCGGGGAAGAAACCTTCAAGGCCAGTCAACAACTATATCCTGTACAGAATGGAGTACGGCATGCGGGAGATAATA TGGTCAACTATCGAGAAGTGCTGGTCGAGGGACGTGTCCACCAGACCATCCCTGGACAGGATCTTGAGTCTCGAG GTCTTCCAGGACGCTGTTCGGAAAGACCTCAGGCCCAATGGCCACTTAAGCTGGTTGTCGACATCTGAGTTCCGC CATGCGATTCGATTTACAGAGAATGACTTCCTTTGA |
| Length | 2211 |
Transcript
| Sequence id | CopciAB_404912.T0 |
|---|---|
| Sequence |
>CopciAB_404912.T0 ATCAACAGACCGCATCTCCAACAGACGACTGACATCCCTTTGCCCCTTGCCCCCACCAAGCATGAGAGAGTTTCA GACAACGAAGAAGCGTCTCAATATTGTCTTCCTTGGTGGCCGTCGGGTTGGGAAGACATCGTTGTTGTCGCAATG GTCTCAGGGAGAATTCCCGAGTGCATATTATCCGACGTTCGAGAACACTGTGGGGAAGGAGGTGACATTTGGGAA CCGAATCTACGACTGTAAGGTCACTGATACAGCGGGCCATGATCGATATACCCCGCTGTCTCCTACCCATGTTGT TGGTATGCACGTATTTGTGCTGGTTTACTCGTTTGAAGACGCCGGATCGTTGGAATTGGTCTCGGCTCTCTACGA GAAGATCTTGGAATTTTTAGGGTCCACCAGACATGCCCGAGTTATCATGGCTGGGAACAAGGCCGATACATCTCG ACGAGAGATATCCTATCCAGAGGCCCACCAACTCGCACAGAAACTGGGCTGCCCTCATCTAATATTGAGTAGCTA CTGGTGGCAAAGTATCGATCAACTATTCGAAGAGTGTACTATAGGCGGTGAACGGAGAAACCGCCAGCCTCCAAC CCGCACATCAGCAACAGCCAAGCCTCAATCTCGTGGCTTGTGGTTCCCAAGCATCGACTTACTATCTTCCCTGCT TCGCTACGCTTCTCGCGTCACCTGGCCCAGCTCCGCGGCAAACACCACCTCAATACCAGAGCCCACTGACACCTA CCATCAGATTCAACGTCACACCAAGCCACACAGCGAGGAGGAATTGCATGGCTCACAGGCCCCCATCCCAATTAC GACGCCATTGGATTACGATCGTCCAACGCGTAGTTCAGTCAGGGGCTACTCAATAATGTCAACCTCATCCGGCGA AGCCACCCTCGTATCTACGTTTGGTCCAAAGTCACGCCTATCGGCCTCATCCAGCAAGGGAGTTAGTCCATTCCA GAAACGGCGCTCTTTTAGCCCCATGACTCCGATCAAGCTCAATGTCACCAAGGACGATGTCGTCAAGAGCAAATT TCGGTTCGTTGATGAACTAACGAGGATACAGAGGGTGCGGGGTGTTGACGCGGGACTTGCACAAGACTTTACGAA CCTATTTCGCTCGATTATGGAAGATAAAGTGACGCTGAAGACCTTGCTGGAATTTCGGGGACATGATGCTCAGGT GGTTCTGAACGCTCTGCAATGGGTTCTGGATTCGCGGAAAGCTCAGCCGGCGGATAAAGGAATGTTGCTGAAGCT GCTTCTTCGACTGTCGAAGCATTCTGGGTCCTATCCTCCGTGTCTGGCGCTGAAAGGAATTAAGCGCGAAGACGA CTATCCGGTTACATCAGGACACTTTGGTGAGGTTTGGAGGGGTCGATACCAGAAGCAGAAAGTTTGCTTGAAGGT GGTCAGGGTGTATCAGCGATCGGATGTTGAGCACTTCCTTCGGATATTCTCTCGCGAAGCTGTGATCTGGAGTCA ACTGAAGCACGACAATGTTCTTCCTTTCTATGGCGTCTATGAACTCGACGAGTCTCGTCGACTTTGCCTGGTCTC TCCTTGGATGGATAACGGAAGTCTTCGGGAGTATGTGCGAAAGAGCTTGTTGTCGGAGGTCCACCATCGCCTTCT GGCAGTCGATGTAACACAGGGCCTGTCCTATCTTCATCAGCAGGGAATGGTCCATGGCGATCTTAAAGGGGATAA CATCCTCATTGACAGTTCTGGACGTGCCTGCATAGCGGACTTCGGGCTCTCAAGTCTGGTGGACGGCGAGATGCT TCGTTGGACAACATCCGAGAGTACCGGACTAGGCGTTGGTGGCACATCCCGCTGGCAAGCTCCAGAACTGTTCGA TCCCGATAATGAATCGAGTCGGCCCACGTGTTATACGGATGTTTATTCGCTTGGTTGCGTTTTCTACGAGATCTT CACTGGAAACATCCCATTCCACGAGACAACACGCGAAGCCACAGTGATATCGTACGTCATGGCGGGGAAGAAACC TTCAAGGCCAGTCAACAACTATATCCTGTACAGAATGGAGTACGGCATGCGGGAGATAATATGGTCAACTATCGA GAAGTGCTGGTCGAGGGACGTGTCCACCAGACCATCCCTGGACAGGATCTTGAGTCTCGAGGTCTTCCAGGACGC TGTTCGGAAAGACCTCAGGCCCAATGGCCACTTAAGCTGGTTGTCGACATCTGAGTTCCGCCATGCGATTCGATT TACAGAGAATGACTTCCTTTGAAGGATCGATTGTAATTTAAAGAAGTACGCGTATTATTTTTAATGTAAACTCTG AACTAAATAGTGT |
| Length | 2338 |
Gene
| Sequence id | CopciAB_404912.T0 |
|---|---|
| Sequence |
>CopciAB_404912.T0 ATCAACAGACCGCATCTCCAACAGACGACTGACATCCCTTTGCCCCTTGCCCCCACCAAGCATGAGAGAGTTTCA GACAACGAAGAAGCGTCTCAATATTGTCTTCCTTGGTGGCCGTCGGGTTGGTGAGTCGAGTTTCTGTGTCTGTCG AGCAGGCCTGGATGTACTCACTCCTACCATCCAGGGAAGACATCGTTGTTGTCGCAATGGTCTCAGGTAGGTACC AAAGTTGCTGTTCGAAGGCGCCAGGCGTTTATTTATATCTCTGATCCACTCCGTTTCAGGGAGAATTCCCGAGTG CATATTATCCGACGTTCGAGAACACTGTGGGGAAGGAGGTGACATTTGGGAACCGAATCTACGACTGTAAGGTCA CTGATACAGCGGGCCATGTACGTTCAATCATGGCTTCCTGGCGCTTCGCTGACTAGATGCACCACACAGGATCGA TATACCCCGCTGTCTCCTACCCATGTTGTTGGTATGCACGTATTTGTGCTGGTTTACTCGTTTGAAGACGCCGGA TCGTTGGAATTGGTCTCGGCTCTCTACGAGAAGATCTTGGAATTTTTAGGGTCCACCAGACATGCCCGAGTTATC ATGGCTGGGAACAAGGCCGATACATCTCGGTAAGTGCTTTGTATCCTATCGTGTCGTGACCGATACTCAACGCTG AACGCCCACAGACGAGAGATATCCTATCCAGAGGCCCACCAACTCGCACAGAAACTGGGCTGCCCTCATCTAATA TTGAGTAGCTACTGGTGGCAAAGTATCGGTGCGTGTACTCGTGGACCCTTCGACTGGTCGTATGCTGATCTCTCA CTTTTACACAGATCAACTATTCGAAGAGTGTACTATAGGCGGTGAACGGAGAAACCGCCAGCCTCCAACCCGCAC ATCAGCAACAGCCAAGCCTCAATCTCGTGGCTTGTGGTTCCCAAGCATCGACTTACTATCTTCCCTGCTTCGCTA CGCTTCTCGCGTCACCTGGCCCAGCTCCGCGGCAAACACCACCTCAATACCAGAGCCCACTGACACCTACCATCA GATTCAACGTCACACCAAGCCACACAGCGAGGAGGAATTGCATGGCTCACAGGCCCCCATCCCAATTACGACGCC ATTGGATTACGATCGTCCAACGCGTAGTTCAGTCAGGGGCTACTCAATAATGTCAACCTCATCCGGCGAAGCCAC CCTCGTATCTACGTTTGGTCCAAAGTCACGCCTATCGGCCTCATCCAGCAAGGGAGTTAGTCCATTCCAGAAACG GCGCTCTTTTAGCCCCATGACTCCGATCAAGCTCAATGTCACCAAGGACGATGTCGTCAAGAGCAAATTTCGGTT CGTTGATGAACTAACGAGGATACAGAGGGTGCGGGGTGTTGACGCGGGACTTGCACAAGACTTTACGAACCTATT TCGCTCGATTATGGAAGATAAAGTGACGCTGAAGACCTTGCTGGAATTTCGGGGACATGATGCTCAGGTGGTTCT GAACGCTCTGCAATGGGTACGTATCATCTCTCTTCTTGGTCGACTAAACTGATTGAGCCTATCCTTAGGTTCTGG ATTCGCGGAAAGCTCAGCCGGCGGATAAAGGAATGTTGCTGAAGCTGCTTCTTCGACTGTCGAAGCATTCTGGGT CCTATCCTCCGTGTCTGGCGCTGAAAGGAATTAAGCGCGAAGACGACTATCCGGTTACATCAGGACACTTTGGTG AGGTTTGGAGGGGTCGATACCAGAAGCAGAAAGTTTGCTTGAAGGTGGTCAGGGTGTATCAGCGATCGGATGTTG AGCACTTCCTTCGGGTACGTTGGAGACTGAAGTTCACAATCGCTATTGGAGCCTGGTTTCACATTCGATTTACTG ACGGGTGAATTTGGCAGATATTCTCTCGCGAAGCTGTGATCTGGAGTCAACTGAAGCACGACAATGTTCTTCCTT TCTATGGCGTCTATGAACTCGACGAGTCTCGTCGACTTTGCCTGGTCTCTCCTTGGATGGATAACGGAAGTCTTC GGGAGTATGTGCGAAAGAGCTTGTTGTCGGAGGTCCACCATCGCCTTCTGGTATGCTTGATACTTCTTTCTTACG CTATAGCGGGGCGCTGACCTTTCATAGGCAGTCGATGTAACACAGGGCCTGTCCTATCTTCATCAGCAGGGAATG GTCCATGGCGATCTTAAAGGGGTACGCTCTTGTCAATGGGCGCTGGATCTCCATAGGAAGTGCTGACGATACTCT TAAGGATAACATCCTCATTGACAGTTCTGGACGTGCCTGCATAGCGGACTTCGGGCTCTCAAGTCTGGTGGACGG CGAGATGCTTCGTTGGACAACATCCGAGAGTACCGGACTAGGCGTTGGTGGCACATCCCGCTGGCAAGCTCCAGA ACTGTTCGATCCCGATAATGAATCGAGTCGGCCCACGTGTTATACGGATGTTTATTCGCTTGGTTGCGTTTTCTA CGAGGTAAATAATTGACCTCAACCCATTCAATGAAACCAAATTGACCAAGGAGATGGACAGATCTTCACTGGAAA CATCCCATTCCACGAGACAACACGCGAAGCCACAGTGATATCGTACGTCATGGCGGGGAAGAAACCTTCAAGGCC AGTCAACAACTATATCCTGTACAGAATGGAGTACGGCATGCGGGAGATAATATGGTCAACTATCGAGAAGTGCTG GTCGAGGGACGTGTCCACCAGACCATCCCTGGACAGGATCTTGAGTCTCGAGGTCTTCCAGGACGCTGTTCGGAA AGACCTCAGGCCCAATGGCCACTTAAGCTGGTTGTCGACATCTGAGTTCCGCCATGCGATTCGATTTACAGAGAA TGACTTCCTTTGAAGGATCGATTGTAATTTAAAGAAGTACGCGTATTATTTTTAATGTAAACTCTGAACTAAATA GTGT |
| Length | 2929 |