CopciAB_413694
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_413694 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 413694 |
| Uniprot id | Functional description | Extension to Ser/Thr-type protein kinases | |
| Location | scaffold_4:1869815..1873756 | Strand | - |
| Gene length (nt) | 3942 | Transcript length (nt) | 2992 |
| CDS length (nt) | 2553 | Protein length (aa) | 850 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN10485_stk21 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7261665 | 78.5 | 0 | 1329 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25182 | 77.4 | 0 | 1287 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1104424 | 72.7 | 0 | 1266 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_71210 | 71.3 | 0 | 1256 |
| Pleurotus ostreatus PC9 | PleosPC9_1_94618 | 73.8 | 0 | 1253 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1500913 | 70.4 | 0 | 1222 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_116050 | 67 | 0 | 1216 |
| Schizophyllum commune H4-8 | Schco3_2632360 | 65.8 | 0 | 1195 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_16334 | 67.9 | 0 | 1179 |
| Lentinula edodes NBRC 111202 | Lenedo1_1047022 | 68.5 | 0 | 1159 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_185181 | 67 | 0 | 1156 |
| Auricularia subglabra | Aurde3_1_1274372 | 66.4 | 0 | 1149 |
| Grifola frondosa | Grifr_OBZ68200 | 70.2 | 0 | 1120 |
| Flammulina velutipes | Flave_chr07AA00572 | 65.2 | 4.941E-304 | 999 |
| Lentinula edodes B17 | Lened_B_1_1_2726 | 76.7 | 1.224E-133 | 447 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_413694.T0 |
| Description | Extension to Ser/Thr-type protein kinases |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd21742 | MobB_NDR_LATS-like | - | 300 | 361 |
| CDD | cd05573 | STKc_ROCK_NDR_like | - | 366 | 801 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 368 | 525 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 588 | 681 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017441 | Protein kinase, ATP binding site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000961 | AGC-kinase, C-terminal |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K00924 |
EggNOG
| COG category | Description |
|---|---|
| T | Extension to Ser/Thr-type protein kinases |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
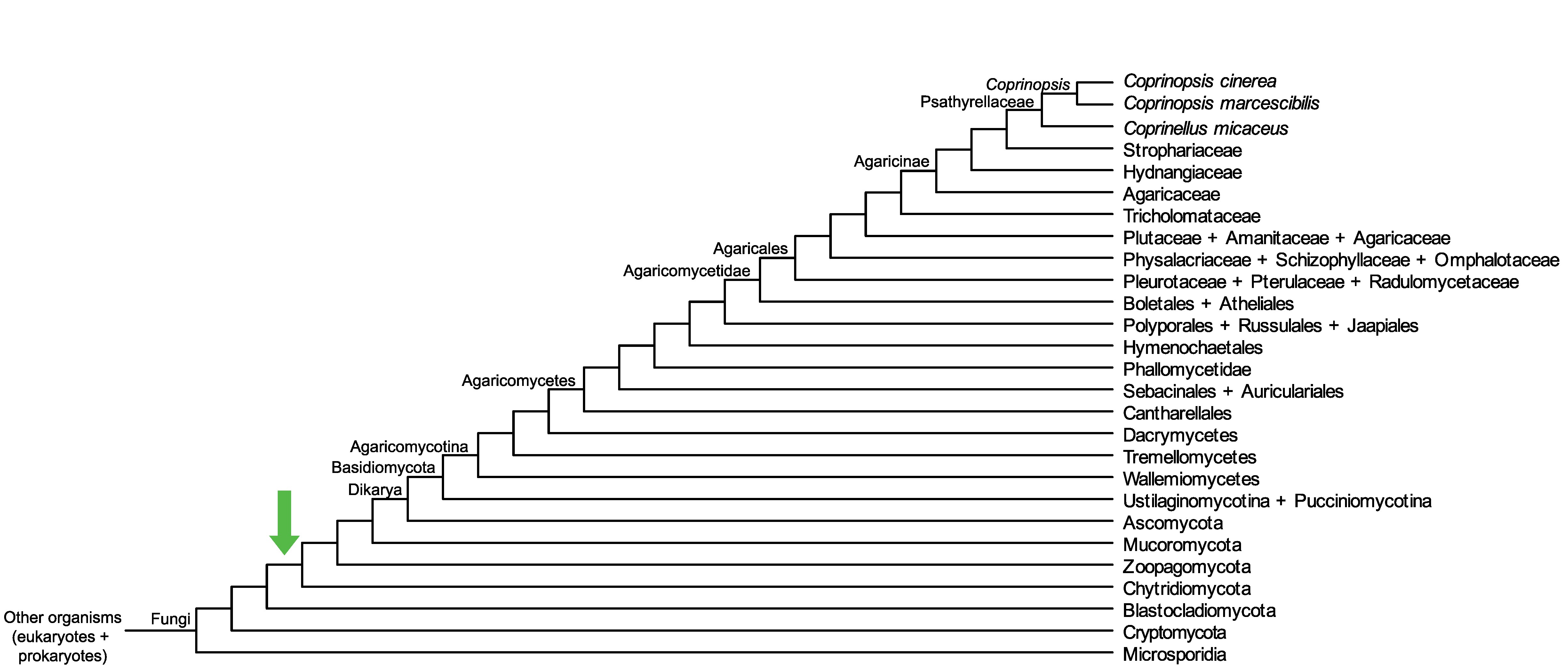
Conservation of CopciAB_413694 across fungi.
Arrow shows the origin of gene family containing CopciAB_413694.
Protein
| Sequence id | CopciAB_413694.T0 |
|---|---|
| Sequence |
>CopciAB_413694.T0 MSSRPLPNPPFQYPQNYWGPVYGDSRNNHHYQDQYHNPYGYPNYAHQPPDFDEQPATSSIPVGTLLHKGFYDLLA MIPTPSPSRLIWGSPPSPNKSSDLQAGPRYEEIVPPANAAIKATPKKGRRVSKDMVSKPTGFVHLVHASDADQYE ALLTRWGPDGIGKLGDPQWANPIKNRVRMLNQAKAVNEVVNACKPSQGSVYGDTTFGQLHVVNGISSATSSSLTT GAIENMSIGPYRYDGLPQRWGGGSTLRKATGGLQTHPEVYEGQDRGASPVMDLPPPPPAKPIIPSLTTLEKAVAA RIYFENIYFSLFRHVPSREQRRLAMERDMAAMGLDSAQKEILRARWRQNETEYLRQTRQKLDASAFIKLKTIGHG AFGVVSLVKEKSSGNLYAMKQMRKADMLRKGQEGHVRAERDILKSASLVHSPGGAEWIVRLHYSFQDRDNLYLIL EYMGGGDLLNLLIERDIFEEDFTRFYVAEMILAIEQCHKHGFIHRDIKPDNFLFDPEGHIKLSDFGLATDLHWAH DTSYYEQQRLHLLHKYGIDLEDPNGIADGRKTKRLDPKQIELLMGGGDGKTGIFTWREKNKRKLAYSICGTNSYM SPEVIRGHGYTYSCDWWSLGVIMFECLYGYPPFVSNSRHVTRQKILNWKQSLRFPSRPRVSPEGVNLMQQLLCEP EDRLGSQASSSVFRPDSLVVQARRSGFMPQAGSTTSVDGAHLIKAHPWFKGIDWDNIHKYPAPYRPELTSPDDTR HFDSDIPPEPLAPANGAPPDATRDPMLKDKVHGEDILNVRKALAFAGFTHKSPRAIEYVRVDRAFDPMPTPRGGG HEDALRGRSPVRETRTIGKGRAISL |
| Length | 850 |
Coding
| Sequence id | CopciAB_413694.T0 |
|---|---|
| Sequence |
>CopciAB_413694.T0 ATGTCCTCGAGACCACTACCGAATCCCCCTTTTCAATATCCTCAGAATTACTGGGGACCGGTCTATGGCGACTCT AGGAATAATCATCACTACCAGGACCAATACCACAACCCCTACGGATACCCCAATTATGCCCACCAGCCTCCTGAC TTCGATGAGCAACCTGCGACGTCTTCCATACCCGTCGGGACCCTTCTCCACAAAGGGTTTTACGATCTCCTCGCG ATGATTCCAACCCCTTCTCCATCCCGGCTCATTTGGGGGAGTCCTCCTAGTCCCAATAAGAGCTCAGATCTGCAA GCTGGTCCCCGATACGAGGAAATAGTACCTCCAGCAAACGCTGCGATCAAGGCGACGCCAAAGAAAGGGCGGAGA GTTAGCAAAGACATGGTTTCAAAACCTACGGGCTTTGTCCACCTAGTACATGCGTCAGACGCGGACCAATATGAA GCTCTCCTAACCAGGTGGGGTCCGGATGGAATCGGCAAACTGGGAGATCCGCAGTGGGCAAACCCGATTAAGAAC CGTGTAAGGATGCTGAACCAAGCTAAGGCTGTCAACGAGGTTGTGAATGCCTGCAAGCCCTCCCAAGGCAGTGTT TATGGAGATACCACTTTTGGACAACTACATGTTGTCAATGGTATCAGCAGCGCGACATCTTCGTCGCTTACCACG GGCGCCATAGAGAATATGAGCATAGGTCCCTACAGATACGACGGTCTTCCGCAACGCTGGGGTGGAGGCTCTACT TTGCGTAAGGCTACGGGAGGGTTGCAAACGCACCCCGAGGTATACGAAGGCCAGGATCGGGGCGCTTCTCCCGTT ATGGACCTTCCTCCGCCGCCCCCTGCCAAACCGATTATACCGTCACTGACAACACTGGAGAAGGCAGTGGCAGCC AGGATTTACTTCGAGAACATCTATTTCTCCCTTTTCCGCCACGTCCCGTCACGCGAACAGAGACGCCTTGCGATG GAACGAGACATGGCAGCGATGGGTCTTGATTCTGCACAGAAGGAGATATTGAGGGCACGTTGGCGGCAGAACGAG ACAGAATACCTCCGCCAGACAAGGCAAAAGTTGGATGCAAGCGCTTTTATCAAACTGAAGACGATAGGCCATGGT GCCTTTGGGGTCGTTTCTCTAGTCAAGGAGAAATCCAGTGGAAATCTTTACGCTATGAAGCAGATGCGCAAGGCA GATATGCTACGCAAGGGTCAAGAGGGGCATGTGCGGGCAGAACGTGACATCTTGAAGTCTGCTTCTCTCGTCCAT TCCCCGGGGGGCGCCGAGTGGATTGTGAGACTCCATTACAGCTTCCAAGACCGGGATAATCTTTATCTGATCCTG GAGTACATGGGCGGTGGAGATCTCCTGAATTTGCTGATAGAACGAGATATATTCGAGGAGGACTTCACCCGATTC TATGTCGCCGAGATGATACTCGCAATCGAACAGTGCCACAAACACGGGTTTATCCACCGCGATATTAAGCCAGAC AATTTCTTGTTCGACCCAGAGGGCCATATCAAATTGAGTGACTTTGGTTTGGCGACCGATCTCCATTGGGCCCAC GATACGTCTTACTACGAACAACAACGCCTGCACCTACTGCACAAATACGGCATTGATCTCGAGGACCCCAATGGC ATTGCTGACGGTCGGAAAACAAAGAGACTAGACCCAAAGCAGATTGAATTGCTCATGGGAGGAGGAGACGGCAAG ACGGGGATCTTCACTTGGCGTGAAAAGAACAAACGCAAGCTGGCATATTCTATTTGCGGCACCAATTCATACATG TCACCAGAAGTGATTAGAGGACATGGTTACACCTATTCGTGTGACTGGTGGAGTTTGGGTGTCATCATGTTCGAG TGTCTCTACGGTTACCCTCCGTTCGTCAGCAACTCCCGACACGTCACCAGACAGAAAATCTTGAACTGGAAGCAG TCTCTCCGTTTTCCTTCCCGTCCCAGAGTTTCACCTGAAGGCGTCAATCTCATGCAGCAGTTGCTCTGTGAACCT GAGGATCGATTGGGATCTCAGGCCTCGTCCTCTGTTTTCAGGCCCGATTCCCTGGTCGTCCAAGCAAGGCGCAGC GGTTTCATGCCTCAGGCTGGTTCAACCACTAGCGTTGACGGTGCCCATCTCATTAAAGCTCATCCGTGGTTCAAA GGGATCGATTGGGACAACATCCACAAGTACCCTGCACCTTACCGCCCGGAACTCACCAGCCCTGACGACACGCGC CATTTCGATTCCGATATCCCACCTGAGCCACTGGCACCTGCCAACGGTGCCCCTCCCGATGCAACGAGGGATCCT ATGTTGAAGGACAAGGTCCACGGAGAAGACATTCTGAACGTCCGCAAAGCCCTCGCTTTCGCTGGATTCACTCAT AAAAGCCCAAGGGCTATCGAATACGTCAGGGTCGACCGAGCATTTGACCCTATGCCCACACCACGAGGCGGCGGC CATGAAGATGCTCTAAGGGGTCGCTCTCCTGTTCGAGAAACACGCACTATTGGCAAGGGTCGGGCGATCTCGTTA TAA |
| Length | 2553 |
Transcript
| Sequence id | CopciAB_413694.T0 |
|---|---|
| Sequence |
>CopciAB_413694.T0 AGGGTCTTCCTCAACCCACCAGCATGTCCTCGAGACCACTACCGAATCCCCCTTTTCAATATCCTCAGAATTACT GGGGACCGGTCTATGGCGACTCTAGGAATAATCATCACTACCAGGACCAATACCACAACCCCTACGGATACCCCA ATTATGCCCACCAGCCTCCTGACTTCGATGAGCAACCTGCGACGTCTTCCATACCCGTCGGGACCCTTCTCCACA AAGGGTTTTACGATCTCCTCGCGATGATTCCAACCCCTTCTCCATCCCGGCTCATTTGGGGGAGTCCTCCTAGTC CCAATAAGAGCTCAGATCTGCAAGCTGGTCCCCGATACGAGGAAATAGTACCTCCAGCAAACGCTGCGATCAAGG CGACGCCAAAGAAAGGGCGGAGAGTTAGCAAAGACATGGTTTCAAAACCTACGGGCTTTGTCCACCTAGTACATG CGTCAGACGCGGACCAATATGAAGCTCTCCTAACCAGGTGGGGTCCGGATGGAATCGGCAAACTGGGAGATCCGC AGTGGGCAAACCCGATTAAGAACCGTGTAAGGATGCTGAACCAAGCTAAGGCTGTCAACGAGGTTGTGAATGCCT GCAAGCCCTCCCAAGGCAGTGTTTATGGAGATACCACTTTTGGACAACTACATGTTGTCAATGGTATCAGCAGCG CGACATCTTCGTCGCTTACCACGGGCGCCATAGAGAATATGAGCATAGGTCCCTACAGATACGACGGTCTTCCGC AACGCTGGGGTGGAGGCTCTACTTTGCGTAAGGCTACGGGAGGGTTGCAAACGCACCCCGAGGTATACGAAGGCC AGGATCGGGGCGCTTCTCCCGTTATGGACCTTCCTCCGCCGCCCCCTGCCAAACCGATTATACCGTCACTGACAA CACTGGAGAAGGCAGTGGCAGCCAGGATTTACTTCGAGAACATCTATTTCTCCCTTTTCCGCCACGTCCCGTCAC GCGAACAGAGACGCCTTGCGATGGAACGAGACATGGCAGCGATGGGTCTTGATTCTGCACAGAAGGAGATATTGA GGGCACGTTGGCGGCAGAACGAGACAGAATACCTCCGCCAGACAAGGCAAAAGTTGGATGCAAGCGCTTTTATCA AACTGAAGACGATAGGCCATGGTGCCTTTGGGGTCGTTTCTCTAGTCAAGGAGAAATCCAGTGGAAATCTTTACG CTATGAAGCAGATGCGCAAGGCAGATATGCTACGCAAGGGTCAAGAGGGGCATGTGCGGGCAGAACGTGACATCT TGAAGTCTGCTTCTCTCGTCCATTCCCCGGGGGGCGCCGAGTGGATTGTGAGACTCCATTACAGCTTCCAAGACC GGGATAATCTTTATCTGATCCTGGAGTACATGGGCGGTGGAGATCTCCTGAATTTGCTGATAGAACGAGATATAT TCGAGGAGGACTTCACCCGATTCTATGTCGCCGAGATGATACTCGCAATCGAACAGTGCCACAAACACGGGTTTA TCCACCGCGATATTAAGCCAGACAATTTCTTGTTCGACCCAGAGGGCCATATCAAATTGAGTGACTTTGGTTTGG CGACCGATCTCCATTGGGCCCACGATACGTCTTACTACGAACAACAACGCCTGCACCTACTGCACAAATACGGCA TTGATCTCGAGGACCCCAATGGCATTGCTGACGGTCGGAAAACAAAGAGACTAGACCCAAAGCAGATTGAATTGC TCATGGGAGGAGGAGACGGCAAGACGGGGATCTTCACTTGGCGTGAAAAGAACAAACGCAAGCTGGCATATTCTA TTTGCGGCACCAATTCATACATGTCACCAGAAGTGATTAGAGGACATGGTTACACCTATTCGTGTGACTGGTGGA GTTTGGGTGTCATCATGTTCGAGTGTCTCTACGGTTACCCTCCGTTCGTCAGCAACTCCCGACACGTCACCAGAC AGAAAATCTTGAACTGGAAGCAGTCTCTCCGTTTTCCTTCCCGTCCCAGAGTTTCACCTGAAGGCGTCAATCTCA TGCAGCAGTTGCTCTGTGAACCTGAGGATCGATTGGGATCTCAGGCCTCGTCCTCTGTTTTCAGGCCCGATTCCC TGGTCGTCCAAGCAAGGCGCAGCGGTTTCATGCCTCAGGCTGGTTCAACCACTAGCGTTGACGGTGCCCATCTCA TTAAAGCTCATCCGTGGTTCAAAGGGATCGATTGGGACAACATCCACAAGTACCCTGCACCTTACCGCCCGGAAC TCACCAGCCCTGACGACACGCGCCATTTCGATTCCGATATCCCACCTGAGCCACTGGCACCTGCCAACGGTGCCC CTCCCGATGCAACGAGGGATCCTATGTTGAAGGACAAGGTCCACGGAGAAGACATTCTGAACGTCCGCAAAGCCC TCGCTTTCGCTGGATTCACTCATAAAAGCCCAAGGGCTATCGAATACGTCAGGGTCGACCGAGCATTTGACCCTA TGCCCACACCACGAGGCGGCGGCCATGAAGATGCTCTAAGGGGTCGCTCTCCTGTTCGAGAAACACGCACTATTG GCAAGGGTCGGGCGATCTCGTTATAATGCGCGGTTGTTTGACTGGTCGACAACACCGCTTCCGCTTGCACCAAGA GCGATCGACTACGATTATGACGGACGGCGCCTAACCTATCAAGGCAAAGACGTCTTTGCTTCTCTGGCCATCTTC CTTTGTATGCAGAGCTCCCTAGCCCTAGCGCTGCGGATCGCCGCTCGTATGCCCGAAAGGTGAGATCATCATGGA GGGGAGGTGTTGCGAGTTTTCCATACCAGTGTTTGTGATGGTCGCACCTATATGTACATGCAAGAGCTGTTCTCG GATGACGGAGGACTTCGATGTGCAACACCCTGTGTGCTTGACATGCTTATATTTTGAATGCTGTATAAATAAGGT TGTGTGTAAGGCTTTACTACTGTAATTGTGGGCGAAAGAATCAATCAGTTAGTGAGCTGTAGACAAA |
| Length | 2992 |
Gene
| Sequence id | CopciAB_413694.T0 |
|---|---|
| Sequence |
>CopciAB_413694.T0 AGGGTCTTCCTCAACCCACCAGCATGTCCTCGAGACCACTACCGAATCCCCCTTTTCAATATCCTCAGAATTACT GGGGACCGGTCTATGGCGACTCTAGGAATAATCATCACTACCAGGACCAATACCACAACCCCTACGGATACCCCA ATTATGCCCACCAGCCTCCTGACTTCGATGAGCAACCTGCGACGTCTTCCATACCCGTCGGGACCCTTCTCCACA AAGGGTTTTACGATCTCCTCGCGATGATTCCAACCCCTTCTCCATCCCGGCTCATTTGGGGGAGTCCTCCTAGTC CCAATAAGAGCTCAGATCTGCAAGCTGGTCCCCGATACGAGGAAATAGTACCTCCAGCAAACGCTGCGATCAAGG CGACGCCAAAGAAAGGGCGGAGAGTTAGCAAAGACATGGTTTCAAAACCTACGGGCTTTGTGTATGTCACGTCGC CCTGTTTCAAACCATACATTCATTTAACAGCCTTTCTTCCCAAATAGCCACCTAGTACATGCGTCAGACGCGGAC CAATATGAAGCTCTCCTAACCAGGTGGGGTCCGGATGGAATCGGCAAACTGGGAGGTCAGTACAGCAGCGCTCCT TCCAATCGCTCCAATGCGAATTACCCTGCCCACAGATCCGCAGTGGGCAAACCCGATTAAGAACCGTGTAAGGAT GCTGAACCAAGCTAAGGCTGTCAACGAGGTTGTGAATGCCTGCAAGCCCTCCCAAGGCAGTGTTTATGGAGATAC CACTTTTGGACAACTACATGTTGTCAATGGTATCAGCAGCGCGACATCTTCGTCGCTTACCACGGGCGCCATAGA GAATATGAGCATAGGTCCCTACAGATACGACGGTCTTCCGCAACGCTGGGGTGGAGGCTCTACTTTGCGTAAGGC TACGGGAGGGTTGCAAACGCACCCCGAGGTATACGAAGGCCAGGATCGGGGCGCTTCTCCCGTTATGGACCTTCC TCCGCCGCCCCCTGCCAAACCGATTATACCGTCACTGACAACACTGGAGAAGGCAGTGGCAGCCAGGATTTACTT CGAGAACATCTATTTCTCCCTTTTCCGCCACGTCCCGTCACGCGAACAGAGACGCCTTGCGATGGAACGAGACAT GGCAGCGATGGGTCTTGATTCTGCACAGAAGGAGATATTGAGGGCACGTTGGCGGCAGAACGAGACAGAATACCT CCGCCAGACAAGGCAAAAGTTGGATGCAAGCGCTTTTATCAAACTGAAGACGATAGGCCATGGTGAGAAGCGCTT CTAGACTATCATTTACCCTGACTCTGATGCCGGAATATCAACAGGTGCCTTTGGGGTCGTTTCTCTAGTCAAGGA GAAATCCAGTGGAAATCTTTACGCTATGAAGCAGGTACCACATGCTCGAAACCCAAATTGACTCGTCGTATTCAA TCCTTTCACAGATGCGCAAGGCAGATATGCTACGCAAGGGTCAAGAGGGGCATGTGCGGGCAGAACGTGACATCT TGAAGTCTGCTTCTCTCGTCCATTCCCCGGGGGGCGCCGAGTGGATTGTGAGACTCCATTACAGCTTCCAAGACC GGGATAATCTTTATCTGGTCAGTCTTATTTGCCTTCCAATGTCGCCGTGTGATTCTTTCTCATATCATTTTCATT TTATCACCCCTGCCTTCTCCCCAACCGGTGTTAATATAGATCCTGGAGTACATGGGCGGTGGAGATCTCCTGAAT TTGCTGATAGAACGAGATATATTCGAGGAGGACTTCACCCGATTCTATGTCGCCGAGGTACATCTGTTTTGTTTA TTTTATCATGACGCCACCTTACTGACCTGGCTGCAGATGATACTCGCAATCGAACAGTGCCACAAACACGGGTTT ATCCACCGCGATATTAAGCCAGACGTGAGCCTCCATCTCACCGCATCGACTGGAGAATTTTAACACTCCATGTTT CACGGGTAGAATTTCTTGTTCGACCCAGAGGGCCATATCAAATTGAGTGACTTTGGTTTGGCGTAAGCACTCCCA TTCCTCTTATTCTGTCTTTCGGTCGGCTCTCAACGACCTTCCATTGCACCAGGACCGATCTCCATTGGGCCCACG ATACGTCTTGTTCGGAATTCCGTCAATTCGCCTCGAACGAATTTTTCCTGACTAGGACTAACAGACTACGAACAA CAACGCCTGCACCTACTGCACAAATACGGCATTGATCTCGAGGACCCCAATGGCATTGCTGACGGTCGGAAAACA AAGAGACTAGACCCAAAGCAGATTGAATTGCTCATGGGAGGAGGAGACGGCAAGACGGGGATCTTCACTTGGCGT GAAAAGAACAAACGCAAGGTATGCTCTCCGCCGCGAAATTTGAGCGTACTTCCTTTCCTGACCATCGGTTCTTGC GAACGTCCAGCTGGCATATTCTATGTAAGATTTGATTATCTGGTCTTCCATAGTTGATTGACTCGGGCCACATTA GTTGCGGCACCAATTCATACAGTAAGGTTTTCTCACCTCAGAAGCAAGGTTACGCCTCACACTCCCCTCTTAGTG TCACCAGAAGTGATTAGAGGACATGGTTACACCTATTCGTGTGACTGGTGGAGTTTGGGTGTCATCATGTTCGAG TGTCTCTACGGGTAGGTCTTCTGCCCCCACGACACTCATGTTCACCGCTAACTTGTCCTTAAGTTACCCTCCGTT CGTCAGCAACTCCGTAGGTGGTCTGAAGTAAATCCAGGACCCAGCATTGATATATGTATGCGCAGCGACACGTCA CCAGACAGAAAATCTTGAACTGGAAGCAGTCTCTCCGTTTTCCTTCCCGTCCCAGAGTTTCACCTGAAGGCGTCA ATCTCATGCAGCAGTTGCTCTGTGAACCTGAGGATCGATTGGGATCTCAGGCCTCGTCCTCTGTTTTCAGGCCCG ATTCCCTGGTCGTCCAAGCAAGGCGCAGCGGTTTCATGCCTCAGGCTGGTTCAACCACTAGCGTTGACGGTGCCC ATCTCATTAAAGTAAGATCCCCGCTTCACTCTCCGTCGAACGTTGCGTCCCGCTCACGCCTTGTGGAACAGGCTC ATCCGTGGTTCAAAGGGATCGATTGGGACAACATCCACAAGTACCCTGCACCTTACCGCCCGGAACTCACCAGCC CTGACGACACGCGCCATTTCGATTCCGATATCCCACCTGAGGTGCGGGCCCAACCTTCCTTATTTCCTCCAACGC CATGATGACCCTGACGGTGTTTCAGCCACTGGCACCTGCCAACGGTGCCCCTCCCGATGCAACGAGGGATCCTAT GTTGAAGGACAAGGTCCACGGAGAAGACATTCTGAACGTCCGCAAAGCCCTCGCTTTCGCTGGATTCACTCATAA AAGCCCAAGGGCTATCGAATACGTCAGGGTCGACCGAGCATTTGACCCTATGCCCACACCACGAGGCGGCGGCCA TGAAGATGCTCTAAGGGGTCGCTCTCCTGTTCGAGAAACACGCACTATTGGCAAGGGTCGGGCGATCTCGTTATA ATGCGCGGTTGTTTGACTGGTCGACAACACCGCTTCCGCTTGCACCAAGAGCGATCGACTACGATTATGACGGAC GGCGCCTAACCTATCAAGGCAAAGACGTCTTTGCTTCTCTGGCCATCTTCCTTTGTATGCAGAGCTCCCTAGCCC TAGCGCTGCGGATCGCCGCTCGTATGCCCGAAAGGTGAGATCATCATGGAGGGGAGGTGTTGCGAGTTTTCCATA CCAGTGTTTGTGATGGTCGCACCTATATGTACATGCAAGAGCTGTTCTCGGATGACGGAGGACTTCGATGTGCAA CACCCTGTGTGCTTGACATGCTTATATTTTGAATGCTGTATAAATAAGGTTGTGTGTAAGGCTTTACTACTGTAA TTGTGGGCGAAAGAATCAATCAGTTAGTGAGCTGTAGACAAA |
| Length | 3942 |