CopciAB_415507
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_415507 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | LKH1 | Synonyms | 415507 |
| Uniprot id | Functional description | Serine/Threonine protein kinases, catalytic domain | |
| Location | scaffold_3:1499487..1502165 | Strand | - |
| Gene length (nt) | 2679 | Transcript length (nt) | 2302 |
| CDS length (nt) | 1611 | Protein length (aa) | 536 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | prk-4 |
| Aspergillus nidulans | AN0988_lkh1 |
| Schizosaccharomyces pombe | lkh1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25581 | 81.9 | 8.748E-292 | 912 |
| Agrocybe aegerita | Agrae_CAA7266941 | 82.3 | 2.065E-296 | 905 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114296 | 77.3 | 8.354E-277 | 848 |
| Flammulina velutipes | Flave_chr09AA01275 | 73.4 | 2.22E-276 | 847 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_134784 | 80.2 | 1.635E-268 | 824 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4068 | 76.2 | 4.865E-268 | 823 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1399302 | 77.7 | 2.659E-266 | 818 |
| Lentinula edodes NBRC 111202 | Lenedo1_1050335 | 75.1 | 1.3E-265 | 816 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1094323 | 77.9 | 2.471E-265 | 815 |
| Grifola frondosa | Grifr_OBZ72301 | 72.1 | 9.139E-260 | 799 |
| Schizophyllum commune H4-8 | Schco3_2503088 | 75.1 | 1.131E-259 | 799 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_132523 | 72.8 | 4.921E-258 | 794 |
| Auricularia subglabra | Aurde3_1_135721 | 72.3 | 5.478E-199 | 624 |
| Pleurotus ostreatus PC9 | PleosPC9_1_86553 | 80.8 | 1.717E-186 | 587 |
| Lentinula edodes B17 | Lened_B_1_1_4315 | 61 | 2.451E-181 | 572 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | LKH1 |
|---|---|
| Protein id | CopciAB_415507.T0 |
| Description | Serine/Threonine protein kinases, catalytic domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14134 | PKc_CLK | - | 189 | 528 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 202 | 528 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08287 |
EggNOG
| COG category | Description |
|---|---|
| T | Serine/Threonine protein kinases, catalytic domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
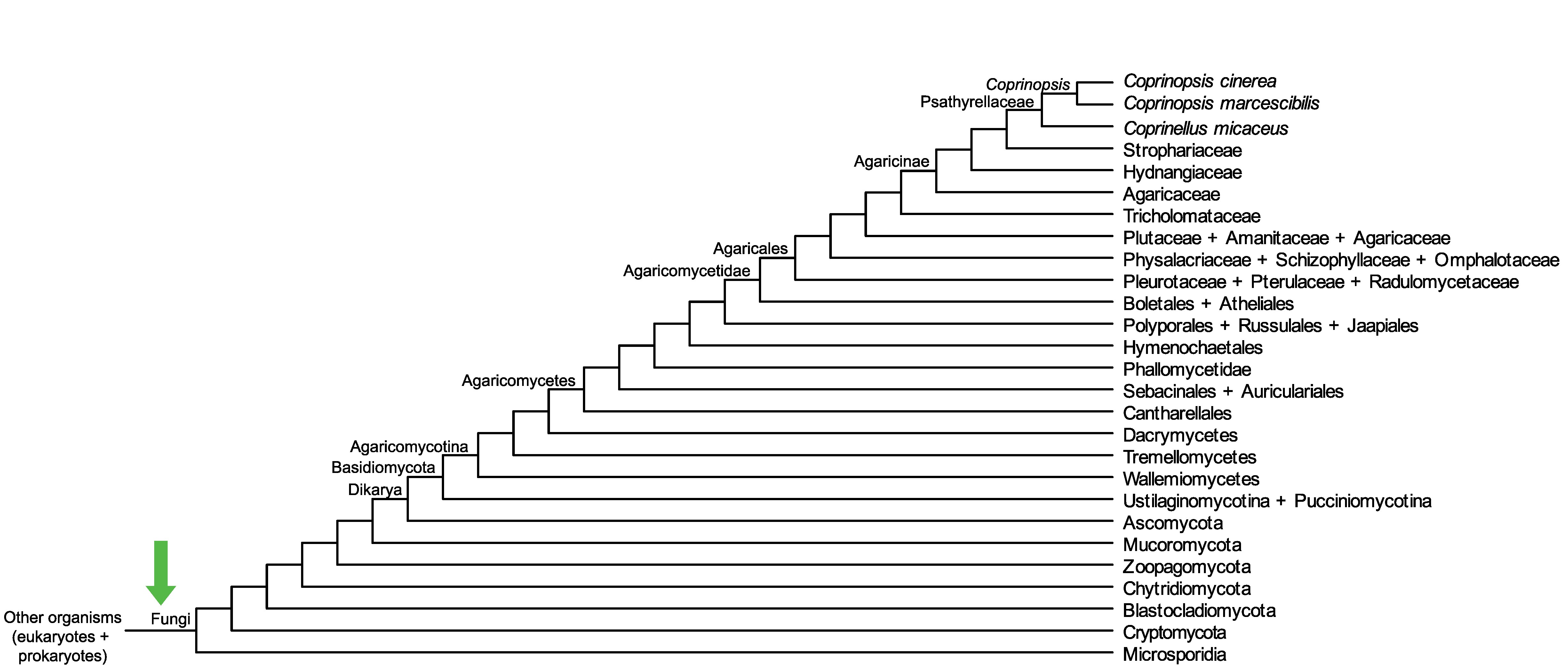
Conservation of CopciAB_415507 across fungi.
Arrow shows the origin of gene family containing CopciAB_415507.
Protein
| Sequence id | CopciAB_415507.T0 |
|---|---|
| Sequence |
>CopciAB_415507.T0 MSAAVHPLNPHHISHYSRSNLPAVTQSIAGPSVMANASPVTTRKRKRAHQYTVSYSEVKELDSDGKVREVIVIED TPPPPTLSPATTHNGGYSASYQPPALAPIRTRARAAAEAQAMSSSSSSVLTAPAPKKRKRDHQDEVRAPPAKKAA AGGQQSNATNAANAWDNRTVAATDDSSKGAPPCDDKEGHYIIVPDDMIFKRYRTVRLLGQGTFGKVVEAIDTQTN NRVAIKIIRAIPKYRDASKIEVRVLQKLKERDPLNRNKCIHLLHWFDHRNHICLVSELLGMCVYDFLKENDFAPF PRHHIQDFARQLFGSVAFLHELRLIHTDLKPENILLVHNDYRTINVPIPGKRNGATKPKKILNSTEIRLIDFGSA TFESEYHSTVVCTRHYRAPEIILGLGWSFPCDAYSLGCILVEFYTGLALYQTHDNLEHLAMMEMVMGKMPERFAR AGARSKPEFFKEGAKLDWPKAKSTRQSKKDVKSTRPLADVIPPYDHINRQFLNLVQKLLAFDPAQRITVREALAH PYFSLTIPEEN |
| Length | 536 |
Coding
| Sequence id | CopciAB_415507.T0 |
|---|---|
| Sequence |
>CopciAB_415507.T0 ATGTCGGCGGCGGTCCACCCGCTCAACCCTCACCATATTTCTCATTATTCTCGTTCAAACTTGCCCGCAGTAACT CAAAGCATCGCTGGGCCCAGCGTCATGGCCAATGCGTCTCCAGTCACAACCAGGAAGCGCAAACGCGCCCACCAA TACACCGTCAGCTACAGCGAAGTCAAGGAGCTCGACAGCGACGGCAAGGTCAGGGAGGTCATCGTCATCGAGGAC ACTCCCCCTCCACCAACTCTCTCACCCGCCACGACACACAACGGTGGTTACTCAGCCTCCTACCAGCCTCCAGCG TTAGCTCCTATCCGCACACGAGCAAGAGCAGCAGCAGAGGCTCAGGCAATGTCTTCGAGCTCTTCGTCCGTCCTG ACAGCCCCCGCTCCAAAGAAGCGAAAGCGGGATCACCAGGATGAGGTCCGTGCGCCCCCTGCAAAGAAAGCCGCT GCTGGTGGCCAACAGTCCAATGCAACCAATGCCGCCAATGCATGGGACAACCGGACCGTTGCTGCTACCGACGAC TCATCTAAAGGCGCTCCTCCATGTGACGACAAGGAGGGCCACTACATCATTGTCCCAGACGACATGATTTTCAAG CGCTATCGCACGGTGCGGCTGCTAGGACAGGGCACCTTCGGTAAGGTGGTGGAAGCCATTGACACACAAACGAAC AACCGCGTTGCTATCAAGATTATCCGGGCCATCCCTAAATACCGCGACGCAAGTAAGATTGAAGTGAGGGTCCTC CAGAAACTCAAGGAGCGTGATCCTCTCAACCGAAACAAATGTATTCACCTCCTACACTGGTTTGACCATCGCAAC CATATATGTCTCGTCTCCGAGCTTCTGGGGATGTGCGTCTACGATTTCTTGAAGGAGAACGACTTTGCTCCCTTC CCCCGGCACCACATCCAAGATTTTGCCAGACAATTGTTCGGCAGTGTCGCATTCCTGCATGAACTCAGGTTGATC CACACCGACTTAAAACCCGAGAATATCCTTCTCGTCCACAATGACTACAGAACCATCAATGTACCTATCCCTGGC AAGCGCAACGGTGCCACAAAACCCAAGAAAATCCTCAACTCGACAGAGATTCGCTTAATCGACTTTGGCTCAGCG ACCTTCGAGTCAGAATACCACTCCACGGTGGTATGCACGCGTCATTACCGTGCACCCGAGATCATTCTTGGTCTT GGTTGGTCGTTCCCCTGTGATGCTTATTCGCTAGGGTGCATCCTCGTCGAGTTCTACACAGGGTTGGCACTGTAC CAAACCCACGACAACCTGGAGCACTTGGCGATGATGGAGATGGTCATGGGCAAAATGCCTGAACGGTTCGCTCGA GCTGGTGCTCGTTCCAAGCCCGAGTTCTTCAAAGAAGGCGCCAAACTCGATTGGCCGAAGGCAAAGTCAACACGA CAAAGCAAGAAAGACGTCAAGTCAACTCGTCCTCTGGCGGATGTCATCCCCCCATATGACCACATCAACCGACAA TTCTTGAACCTGGTGCAGAAGCTACTGGCCTTTGATCCAGCACAGAGGATCACTGTCAGAGAAGCGCTCGCCCAC CCCTACTTCTCTCTGACAATACCGGAAGAAAACTAA |
| Length | 1611 |
Transcript
| Sequence id | CopciAB_415507.T0 |
|---|---|
| Sequence |
>CopciAB_415507.T0 AGTCATTTGCAGCTCACTCATCGCCGTCTCTCTCTCTTCTTCCCTCAATCATCCACGGTCCTTTGCTTCAGCCAT GTCGGCGGCGGTCCACCCGCTCAACCCTCACCATATTTCTCATTATTCTCGTTCAAACTTGCCCGCAGTAACTCA AAGCATCGCTGGGCCCAGCGTCATGGCCAATGCGTCTCCAGTCACAACCAGGAAGCGCAAACGCGCCCACCAATA CACCGTCAGCTACAGCGAAGTCAAGGAGCTCGACAGCGACGGCAAGGTCAGGGAGGTCATCGTCATCGAGGACAC TCCCCCTCCACCAACTCTCTCACCCGCCACGACACACAACGGTGGTTACTCAGCCTCCTACCAGCCTCCAGCGTT AGCTCCTATCCGCACACGAGCAAGAGCAGCAGCAGAGGCTCAGGCAATGTCTTCGAGCTCTTCGTCCGTCCTGAC AGCCCCCGCTCCAAAGAAGCGAAAGCGGGATCACCAGGATGAGGTCCGTGCGCCCCCTGCAAAGAAAGCCGCTGC TGGTGGCCAACAGTCCAATGCAACCAATGCCGCCAATGCATGGGACAACCGGACCGTTGCTGCTACCGACGACTC ATCTAAAGGCGCTCCTCCATGTGACGACAAGGAGGGCCACTACATCATTGTCCCAGACGACATGATTTTCAAGCG CTATCGCACGGTGCGGCTGCTAGGACAGGGCACCTTCGGTAAGGTGGTGGAAGCCATTGACACACAAACGAACAA CCGCGTTGCTATCAAGATTATCCGGGCCATCCCTAAATACCGCGACGCAAGTAAGATTGAAGTGAGGGTCCTCCA GAAACTCAAGGAGCGTGATCCTCTCAACCGAAACAAATGTATTCACCTCCTACACTGGTTTGACCATCGCAACCA TATATGTCTCGTCTCCGAGCTTCTGGGGATGTGCGTCTACGATTTCTTGAAGGAGAACGACTTTGCTCCCTTCCC CCGGCACCACATCCAAGATTTTGCCAGACAATTGTTCGGCAGTGTCGCATTCCTGCATGAACTCAGGTTGATCCA CACCGACTTAAAACCCGAGAATATCCTTCTCGTCCACAATGACTACAGAACCATCAATGTACCTATCCCTGGCAA GCGCAACGGTGCCACAAAACCCAAGAAAATCCTCAACTCGACAGAGATTCGCTTAATCGACTTTGGCTCAGCGAC CTTCGAGTCAGAATACCACTCCACGGTGGTATGCACGCGTCATTACCGTGCACCCGAGATCATTCTTGGTCTTGG TTGGTCGTTCCCCTGTGATGCTTATTCGCTAGGGTGCATCCTCGTCGAGTTCTACACAGGGTTGGCACTGTACCA AACCCACGACAACCTGGAGCACTTGGCGATGATGGAGATGGTCATGGGCAAAATGCCTGAACGGTTCGCTCGAGC TGGTGCTCGTTCCAAGCCCGAGTTCTTCAAAGAAGGCGCCAAACTCGATTGGCCGAAGGCAAAGTCAACACGACA AAGCAAGAAAGACGTCAAGTCAACTCGTCCTCTGGCGGATGTCATCCCCCCATATGACCACATCAACCGACAATT CTTGAACCTGGTGCAGAAGCTACTGGCCTTTGATCCAGCACAGAGGATCACTGTCAGAGAAGCGCTCGCCCACCC CTACTTCTCTCTGACAATACCGGAAGAAAACTAAGCCCGGACTACTTCCTCTATTTTTTGCTTTGCTTTCTCCTC GTTTCCATCGCCTCATCATTGTTCATTGTGCCACTATCAACAGTCAAAGATTCGCAACACTGTAACACAAAGAAG TCACGACGCCCGGCTCCAGGACCACCGACGTCTTGGAAGCATGTCCACCATGGATGACACCTTTTTTTGCCCCAG CTGGCTTGTACTACATTATGCATTCATTTAGATCATCACCTATCAGATGAACAACCGGTCGCTCATTACAACAAA CCCAGTAACCCTCGTTCGTCCCCCGCCTTTCATCTTCCTTCTTGCATCCACTGGCCACATATCTTGCTTAGGGCG GCGCCATTTTCTACAAACCCCCTCGTTCGCGTCTGGTAACAACGAAACGGACTCCTTTGTCCCTTAGCCTTGTAT ACTGTTCCCTCTCAACCCTCGTTTTGTTTTAATATTTCCTTATTCTTATACTTGGAACTCGACCATTCCCCGTTC GTTTCTTCTCTCTATTCACGTTTTTCTCTTCTCTTTCTGGTTGCCTGGATGCGTTTAATCTGATACGATCTCGAC GTTTGTGTATAAATATAGTCAATAATAGAGCGGCCAAGTCCCGGAAACCGTC |
| Length | 2302 |
Gene
| Sequence id | CopciAB_415507.T0 |
|---|---|
| Sequence |
>CopciAB_415507.T0 AGTCATTTGCAGCTCACTCATCGCCGTCTCTCTCTCTTCTTCCCTCAATCATCCACGGTCCTTTGCTTCAGCCAT GTCGGCGGCGGTCCACCCGCTCAACCCTCACCATATTTCTCATTATTCTCGTTCAAACTTGCCCGCAGTAACTCA AAGCATCGCTGGGCCCAGCGTCATGGCCAATGCGTCTCCAGTCACAACCAGGAAGCGCAAACGCGCCCACCAATA CACCGTCAGCTACAGCGAAGTCAAGGAGCTCGACAGCGACGGCAAGGTCAGGGAGGTCATCGTCATCGAGGACAC TCCCCCTCCACCAACTCTCTCACCCGCCACGACACACAACGGTGGTTACTCAGCCTCCTACCAGCCTCCAGCGTT AGCTCCTATCCGCACACGAGCAAGAGCAGCAGCAGAGGCTCAGGCAATGTCTTCGAGCTCTTCGTCCGTCCTGAC AGCCCCCGCTCCAAAGAAGCGAAAGCGGGATCACCAGGATGAGGTCCGTGCGCCCCCTGCAAAGAAAGCCGCTGC TGGTGGCCAACAGTCCAATGCAACCAATGCCGCCAATGCATGGGACAACCGGACCGTTGCTGCTACCGACGACGT AGGTTTCATCTCCTTTTCGCCCGCCTTCGATAACTCTGGGCTAACCTCATCGATCACCGCCCCAAATAGTCATCT AAAGGCGCTCCTCCATGTGACGACAAGGAGGGCCACTACATCATTGTCCCAGACGACATGATTTTCAAGCGCTGT GTGTCCCCCGCCTTTCTTGGCTTCTTCTTATGCTGACGTCTTCGCCTAGATCGCACGGTGCGGCTGCTAGGACAG GGCACCTTCGGTAAGGTGGTGGAAGCCATTGACACACAAACGAACAACCGCGTTGCTATCAAGATTATCCGGGCC ATCCCTAAATACCGCGACGCAAGTAAGATTGAAGTGAGGGTCCTCCAGAAACTCAAGGAGCGTGATCCTCTCAAC CGAAAGTACGTTTTCTACCTCATCGTTTAAGCGGATGGAAGTTAACCCTTGGTCAGCAAATGTATTCACCTCCTA CACTGGTTTGACCATCGCAACCATATATGTCTCGTCTCCGAGCTTCTGGGGATGTGCGTCTACGATTTCTTGAAG GAGAACGACTTTGCTCCCTTCCCCCGGCACCACATCCAAGATTTTGCCAGACAATTGTTCGGCAGTGTCGCATGT GAGATCCGGCCAATCCATCTCCCCATTTGTGCGCCTACTGACTATTTTTTTCTAGTCCTGCATGAACTCAGGTTG ATCCACACCGACTTAAAACCCGAGAATATCCTTCTCGTCCACAATGACTACAGAACCATCAATGTACCTATCCCT GGCAAGGTATGACTTTCGCACCACCTGTTTCGTGCACCTCCCTCATGGTGGAACAGCGCAACGGTGCCACAAAAC CCAAGAAAATCCTCAACTCGACAGAGATTCGCTTAATCGACTTTGGCTCAGCGACCTTCGAGTCAGAATACCACT CCACGGTGGTATGCACGCGTCATTACCGTGCACCCGAGATCATTCTTGGTGTGTTCATTTGCTTCTTACTATCCA TTGTCCACTAAGCCTTTTCGTAGGTCTTGGTTGGTCGTTCCCCTGTGATGCTTATTCGCTAGGGTGCATCCTCGT CGAGTTCTACACAGGGTTGGCACTGTACCAAACCCACGACAACCTGGAGCACTTGGCGATGATGGAGATGGTCAT GGGCAAAATGCCTGAACGGTTCGCTCGAGCTGGTGCTCGTTCCAAGCCCGAGTTCTTCAAAGAAGGCGCCAAACT CGATTGGCCGAAGGCAAAGTCAACACGACAAAGCAAGAAAGACGTCAAGTCAACTCGTCCTCTGGCGGTAAGTAA TTGCTGCGCCGAATGGAGCATTAGCTGACGGCACGACAGGATGTCATCCCCCCATATGACCACATCAACCGACAA TTCTTGAACCTGGTGCAGAAGCTACTGGCCTTTGATCCAGCACAGAGGATCACTGTCAGAGAAGCGCTCGCCCAC CCCTACTTCTCTCTGACAATACCGGAAGAAAACTAAGCCCGGACTACTTCCTCTATTTTTTGCTTTGCTTTCTCC TCGTTTCCATCGCCTCATCATTGTTCATTGTGCCACTATCAACAGTCAAAGATTCGCAACACTGTAACACAAAGA AGTCACGACGCCCGGCTCCAGGACCACCGACGTCTTGGAAGCATGTCCACCATGGATGACACCTTTTTTTGCCCC AGCTGGCTTGTACTACATTATGCATTCATTTAGATCATCACCTATCAGATGAACAACCGGTCGCTCATTACAACA AACCCAGTAACCCTCGTTCGTCCCCCGCCTTTCATCTTCCTTCTTGCATCCACTGGCCACATATCTTGCTTAGGG CGGCGCCATTTTCTACAAACCCCCTCGTTCGCGTCTGGTAACAACGAAACGGACTCCTTTGTCCCTTAGCCTTGT ATACTGTTCCCTCTCAACCCTCGTTTTGTTTTAATATTTCCTTATTCTTATACTTGGAACTCGACCATTCCCCGT TCGTTTCTTCTCTCTATTCACGTTTTTCTCTTCTCTTTCTGGTTGCCTGGATGCGTTTAATCTGATACGATCTCG ACGTTTGTGTATAAATATAGTCAATAATAGAGCGGCCAAGTCCCGGAAACCGTC |
| Length | 2679 |