CopciAB_416523
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_416523 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | FUN31 | Synonyms | 416523 |
| Uniprot id | Functional description | Camk camkl pask protein kinase | |
| Location | scaffold_1:2610042..2612268 | Strand | - |
| Gene length (nt) | 2227 | Transcript length (nt) | 2073 |
| CDS length (nt) | 1761 | Protein length (aa) | 586 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29572 | 66.3 | 3.561E-244 | 757 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1383428 | 65.8 | 8.751E-241 | 747 |
| Pleurotus ostreatus PC15 | PleosPC15_2_38487 | 65.7 | 8.833E-239 | 741 |
| Pleurotus ostreatus PC9 | PleosPC9_1_85662 | 65.7 | 8.646E-239 | 741 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_1296 | 62.9 | 3.786E-232 | 722 |
| Lentinula edodes NBRC 111202 | Lenedo1_1081303 | 62.9 | 1.874E-231 | 720 |
| Schizophyllum commune H4-8 | Schco3_2611791 | 64.9 | 1.138E-229 | 715 |
| Flammulina velutipes | Flave_chr08AA00902 | 64 | 1.299E-225 | 703 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_189661 | 75.2 | 1.589E-220 | 688 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_110440 | 75.2 | 1.655E-220 | 688 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_38204 | 60.5 | 5.166E-218 | 681 |
| Grifola frondosa | Grifr_OBZ79338 | 59.9 | 7.844E-213 | 666 |
| Lentinula edodes B17 | Lened_B_1_1_3413 | 61.6 | 3.611E-211 | 661 |
| Agrocybe aegerita | Agrae_CAA7266481 | 63.4 | 6.949E-187 | 591 |
| Auricularia subglabra | Aurde3_1_1228032 | 58.8 | 1.605E-150 | 486 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | FUN31 |
|---|---|
| Protein id | CopciAB_416523.T0 |
| Description | Camk camkl pask protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 348 | 574 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| K08286 |
EggNOG
| COG category | Description |
|---|---|
| T | Camk camkl pask protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
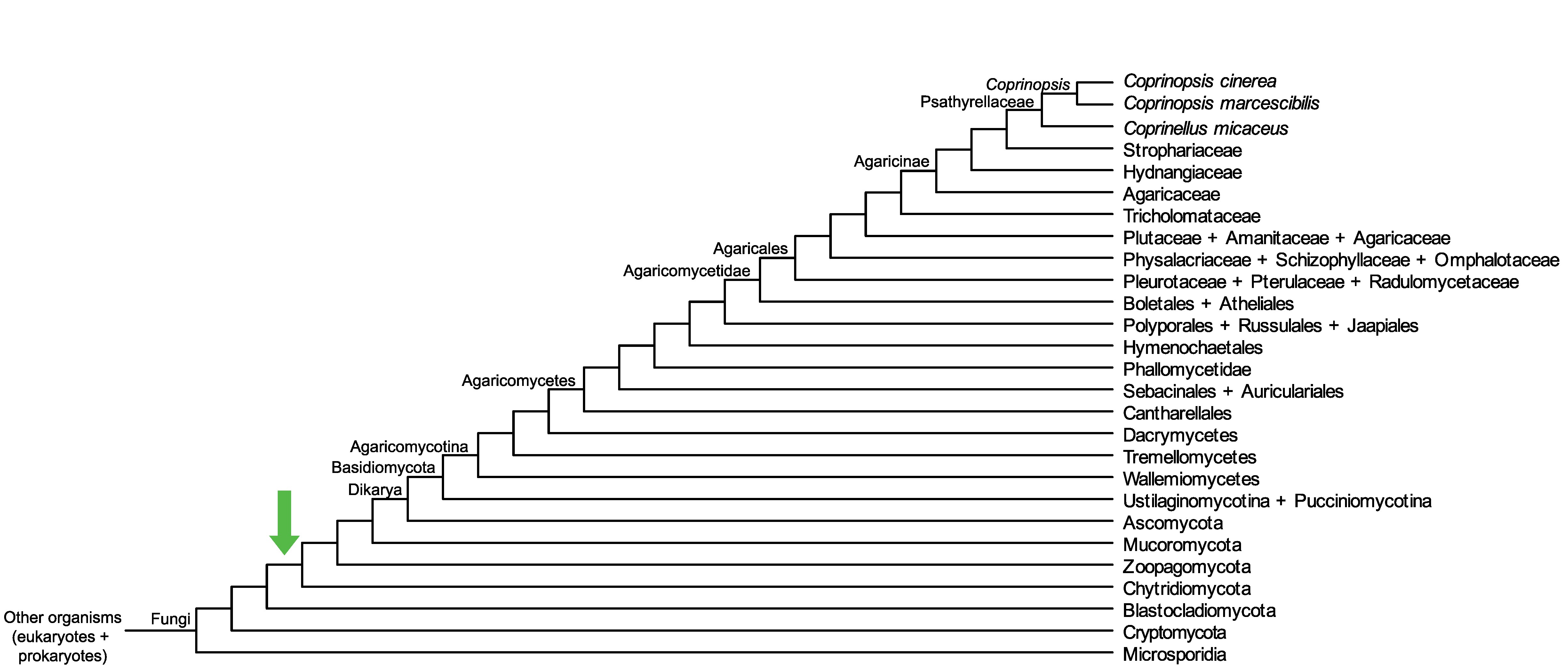
Conservation of CopciAB_416523 across fungi.
Arrow shows the origin of gene family containing CopciAB_416523.
Protein
| Sequence id | CopciAB_416523.T0 |
|---|---|
| Sequence |
>CopciAB_416523.T0 MLSTSSYKLAFDEYDYDSDWTIPTQPPKVYRSASVKSWGVTSIGRGFDWDEEKNSSVNLDEAAAAATSSRQPERA NEAFDSHVRKHHHHHHHHSQPGKSPVPSSVSSSASLTNPKLSQYPSLMAQSHSLPQSRSASPNPLSSAASFASFR SPTSASTSPVPSSPRPRRRSSQQRVSLIAGRVSIAPIEPPPSPPPLLPQSLRRTPSSGSILSTAASTRAPSPASD NQSFIGGRSISEFEIEGEIGRGAYGLVKRAREILSDGSLGPPLIIKQVIKSRILADCWKKHPRHGTIPIEIYVMS AISNTSYILPPKRPWDPSRFSQEKSKDALQLDLVSEQSDDDWVEGKVVKGHPNICPLLDFFEDNHYYYLVLPFTS PDRRPGQTSAASDLFDLVESYPNGLPPFLVRSYLGQIADALAFLHSHGIVHRDVKDENVILGPRDKAILIDFGSS GLIKKTGWDTFSGTLDYAGPEILRGERYYGKEQDVWAFGVVAYVLLVGECPFMTATEAQEGFSSPFANASISLDE RCGEGKEGEGEEPDGGGALGDAAVLVRACLAVDPAARPTFEKIMQSRFLAGKGGWGMEKPM |
| Length | 586 |
Coding
| Sequence id | CopciAB_416523.T0 |
|---|---|
| Sequence |
>CopciAB_416523.T0 ATGCTCTCGACCTCGAGTTATAAGCTGGCCTTCGACGAGTATGATTATGATTCCGATTGGACCATTCCGACGCAG CCACCAAAAGTGTATCGATCCGCCAGCGTGAAGAGCTGGGGCGTGACGTCGATTGGGCGGGGATTCGACTGGGAC GAAGAGAAAAACAGCAGCGTTAATCTGGACGAGGCGGCGGCGGCGGCAACGAGCAGCAGGCAACCAGAAAGGGCG AACGAAGCCTTCGATTCACATGTCCGCAAGCACCACCACCACCACCACCACCATTCGCAGCCAGGCAAATCTCCG GTTCCTTCCTCTGTCTCATCATCTGCCTCTCTCACCAACCCCAAACTTTCCCAGTATCCCAGCCTAATGGCCCAG TCTCACTCTCTGCCCCAGTCCCGTTCCGCCTCCCCCAATCCCTTGTCATCCGCTGCTTCCTTTGCTTCCTTCAGA AGCCCGACCAGTGCTTCGACGTCGCCGGTCCCCTCGTCGCCCCGACCACGTCGCAGATCGTCTCAGCAGCGCGTT TCGCTCATTGCCGGCCGAGTCTCGATAGCGCCCATCGAACCACCTCCCTCGCCCCCGCCCTTGCTGCCCCAGAGC TTGAGGCGCACACCCAGCTCGGGCAGCATCCTGAGCACAGCGGCAAGCACCCGGGCTCCGTCCCCTGCGTCCGAT AACCAGTCCTTCATCGGAGGAAGGAGCATTTCCGAATTTGAAATAGAAGGAGAGATTGGCCGGGGTGCCTATGGA CTCGTCAAGCGGGCGAGAGAGATCCTTTCCGATGGTTCACTTGGGCCACCGCTTATCATCAAGCAAGTCATCAAG TCTCGCATCCTCGCCGATTGCTGGAAGAAACATCCTCGCCATGGCACTATCCCCATCGAAATCTATGTCATGTCC GCCATCTCCAATACTTCCTACATCCTTCCACCAAAGCGACCATGGGACCCTTCCCGCTTCTCGCAAGAAAAATCA AAAGACGCCTTGCAACTTGATCTCGTTTCAGAACAGTCCGACGATGACTGGGTAGAAGGCAAGGTCGTCAAAGGA CATCCTAATATATGCCCCCTCCTCGACTTCTTCGAAGACAATCATTATTACTACTTGGTCCTTCCCTTCACATCC CCAGACCGCAGGCCGGGTCAAACGTCTGCTGCCTCTGATCTCTTCGATCTCGTAGAAAGTTATCCTAACGGCTTG CCCCCATTCCTCGTACGCAGCTACCTCGGGCAAATCGCCGACGCCCTTGCCTTTTTGCACTCCCATGGAATCGTC CACCGTGATGTCAAGGATGAAAACGTTATCTTAGGCCCCCGAGATAAAGCCATCCTAATCGATTTCGGAAGCTCG GGGTTAATCAAGAAAACGGGATGGGATACTTTTAGCGGGACACTGGACTACGCAGGCCCTGAAATTCTCCGGGGC GAGCGCTACTATGGAAAAGAGCAAGATGTGTGGGCATTTGGCGTTGTCGCTTATGTTCTCCTCGTGGGCGAGTGT CCATTCATGACAGCGACCGAAGCACAAGAAGGCTTTTCTTCACCTTTCGCCAATGCCTCCATTTCTCTTGATGAA CGTTGCGGCGAAGGAAAGGAAGGTGAGGGCGAGGAGCCAGATGGAGGTGGAGCTCTTGGCGATGCTGCTGTGCTC GTTCGAGCATGCCTGGCCGTGGATCCCGCTGCGAGACCGACGTTTGAAAAAATCATGCAATCCCGATTCTTGGCT GGAAAGGGCGGATGGGGAATGGAGAAGCCAATGTGA |
| Length | 1761 |
Transcript
| Sequence id | CopciAB_416523.T0 |
|---|---|
| Sequence |
>CopciAB_416523.T0 GAGCATCCCTTGCAGCTGTGAATAAATCATTGGTCGGACCTGGGCATCGCCTCGCACAGCGACCAAGCCTCTTCT TTCTCTACCACCTCCTTTGATGCTCTCGACCTCGAGTTATAAGCTGGCCTTCGACGAGTATGATTATGATTCCGA TTGGACCATTCCGACGCAGCCACCAAAAGTGTATCGATCCGCCAGCGTGAAGAGCTGGGGCGTGACGTCGATTGG GCGGGGATTCGACTGGGACGAAGAGAAAAACAGCAGCGTTAATCTGGACGAGGCGGCGGCGGCGGCAACGAGCAG CAGGCAACCAGAAAGGGCGAACGAAGCCTTCGATTCACATGTCCGCAAGCACCACCACCACCACCACCACCATTC GCAGCCAGGCAAATCTCCGGTTCCTTCCTCTGTCTCATCATCTGCCTCTCTCACCAACCCCAAACTTTCCCAGTA TCCCAGCCTAATGGCCCAGTCTCACTCTCTGCCCCAGTCCCGTTCCGCCTCCCCCAATCCCTTGTCATCCGCTGC TTCCTTTGCTTCCTTCAGAAGCCCGACCAGTGCTTCGACGTCGCCGGTCCCCTCGTCGCCCCGACCACGTCGCAG ATCGTCTCAGCAGCGCGTTTCGCTCATTGCCGGCCGAGTCTCGATAGCGCCCATCGAACCACCTCCCTCGCCCCC GCCCTTGCTGCCCCAGAGCTTGAGGCGCACACCCAGCTCGGGCAGCATCCTGAGCACAGCGGCAAGCACCCGGGC TCCGTCCCCTGCGTCCGATAACCAGTCCTTCATCGGAGGAAGGAGCATTTCCGAATTTGAAATAGAAGGAGAGAT TGGCCGGGGTGCCTATGGACTCGTCAAGCGGGCGAGAGAGATCCTTTCCGATGGTTCACTTGGGCCACCGCTTAT CATCAAGCAAGTCATCAAGTCTCGCATCCTCGCCGATTGCTGGAAGAAACATCCTCGCCATGGCACTATCCCCAT CGAAATCTATGTCATGTCCGCCATCTCCAATACTTCCTACATCCTTCCACCAAAGCGACCATGGGACCCTTCCCG CTTCTCGCAAGAAAAATCAAAAGACGCCTTGCAACTTGATCTCGTTTCAGAACAGTCCGACGATGACTGGGTAGA AGGCAAGGTCGTCAAAGGACATCCTAATATATGCCCCCTCCTCGACTTCTTCGAAGACAATCATTATTACTACTT GGTCCTTCCCTTCACATCCCCAGACCGCAGGCCGGGTCAAACGTCTGCTGCCTCTGATCTCTTCGATCTCGTAGA AAGTTATCCTAACGGCTTGCCCCCATTCCTCGTACGCAGCTACCTCGGGCAAATCGCCGACGCCCTTGCCTTTTT GCACTCCCATGGAATCGTCCACCGTGATGTCAAGGATGAAAACGTTATCTTAGGCCCCCGAGATAAAGCCATCCT AATCGATTTCGGAAGCTCGGGGTTAATCAAGAAAACGGGATGGGATACTTTTAGCGGGACACTGGACTACGCAGG CCCTGAAATTCTCCGGGGCGAGCGCTACTATGGAAAAGAGCAAGATGTGTGGGCATTTGGCGTTGTCGCTTATGT TCTCCTCGTGGGCGAGTGTCCATTCATGACAGCGACCGAAGCACAAGAAGGCTTTTCTTCACCTTTCGCCAATGC CTCCATTTCTCTTGATGAACGTTGCGGCGAAGGAAAGGAAGGTGAGGGCGAGGAGCCAGATGGAGGTGGAGCTCT TGGCGATGCTGCTGTGCTCGTTCGAGCATGCCTGGCCGTGGATCCCGCTGCGAGACCGACGTTTGAAAAAATCAT GCAATCCCGATTCTTGGCTGGAAAGGGCGGATGGGGAATGGAGAAGCCAATGTGATTTTCCCGACATAGTAGACG TCCATTTTCGCACTAGATGAAGGCGATTGGGAGTCAGTCTTGTGACAGTAATGTTATCGATAGTGATTGTGACCG GCGAAATGTACCATCCCGTGTCTGTATTCTGTGATTGAAACTAGCCTATGTAGACTTAGGAAGATGCCAATGTTG TAGAGTAGATATTACAGTGTACCTCTAACTCATTGTCCTATTTATTGC |
| Length | 2073 |
Gene
| Sequence id | CopciAB_416523.T0 |
|---|---|
| Sequence |
>CopciAB_416523.T0 GAGCATCCCTTGCAGCTGTGAATAAATCATTGGTCGGACCTGGGCATCGCCTCGCACAGCGACCAAGCCTCTTCT TTCTCTACCACCTCCTTTGATGCTCTCGACCTCGAGTTATAAGCTGGCCTTCGACGAGTATGATTATGATTCCGA TTGGACCATTCCGACGCAGCCACCAAAAGTGTATCGATCCGCCAGCGTGAAGAGCTGGGGCGTGACGTCGATTGG GCGGGGATTCGACTGGGACGAAGAGAAAAACAGCAGCGTTAATCTGGACGAGGCGGCGGCGGCGGCAACGAGCAG CAGGCAACCAGAAAGGGCGAACGAAGCCTTCGATTCACATGTCCGCAAGCACCACCACCACCACCACCACCATTC GCAGCCAGGCAAATCTCCGGTTCCTTCCTCTGTCTCATCATCTGCCTCTCTCACCAACCCCAAACTTTCCCAGTA TCCCAGCCTAATGGCCCAGTCTCACTCTCTGCCCCAGTCCCGTTCCGCCTCCCCCAATCCCTTGTCATCCGCTGC TTCCTTTGCTTCCTTCAGAAGCCCGACCAGTGCTTCGACGTCGCCGGTCCCCTCGTCGCCCCGACCACGTCGCAG ATCGTCTCAGCAGCGCGTTTCGCTCATTGCCGGCCGAGTCTCGATAGCGCCCATCGAACCACCTCCCTCGCCCCC GCCCTTGCTGCCCCAGAGCTTGAGGCGCACACCCAGCTCGGGCAGCATCCTGAGCACAGCGGCAAGCACCCGGGC TCCGTCCCCTGCGTCCGATAACCAGTCCTTCATCGGAGGAAGGAGCATTTCCGAATTTGAAATAGAAGGAGAGAT TGGCCGGGGTGCCTATGGACTCGTCAAGCGGGCGAGAGAGATCCTTTCCGATGGTTCACTTGGGGTGTGTACATT ATCCTCGTTGCTTAAACGCCATTGATATCCTTGTCTAGCCACCGCTTATCATCAAGCAAGTCATCAAGTCTCGCA TCCTCGCCGATTGCTGGAAGAAACATCCTCGCCATGGCACTATCCCCATCGAAATCTATGTCATGTCCGCCATCT CCAATACTTCCTACATCCTTCCACCAAAGCGACCATGGGACCCTTCCCGCTTCTCGCAAGAAAAATCAAAAGACG CCTTGCAACTTGATCTCGTTTCAGAACAGTCCGACGATGACTGGGTAGAAGGCAAGGTCGTCAAAGGACATCCTA ATATATGCCCCCTCCTCGACTTCTTCGAAGACAATCATTATTACTACTTGGTCCTTCCCTTCACATCCCCAGACC GCAGGCCGGGTCAAACGTCTGCTGCCTCTGATCTCTTCGATCTCGTAGAAAGTTATCCTAACGGCTTGCCCCCAT TCCTCGTACGCAGCTACCTCGGGCAAATCGCCGACGCCCTTGCCTTTTTGCACTCCCATGGAATCGGTGAGTGTC GCCTCCTCATTCTCGATTGATCTGGTTCTGATTTCTCGTCAGTCCACCGTGATGTCAAGGATGAAAACGTTATCT TAGGCCCCCGAGATAAAGCCATCCTAATCGATTTCGGAAGCTCGGGGTTAATCAAGAAAACGGGATGGGATACTT TTAGCGGGACGTGAGTAACTCGTTTCAATGTTATCTATTCACTTCTGATCGGTTCGCTTTGTAGACTGGACTACG CAGGCCCTGAAATTCTCCGGGGCGAGCGCTACTATGGAAAAGAGCAAGATGTGTGGGCATTTGGCGTTGTCGCTT ATGTTCTCCTCGTGGGCGAGTGTCCATTCATGACAGCGACCGAAGCACAAGAAGGCTTTTCTTCACCTTTCGCCA ATGCCTCCATTTCTCTTGATGAACGTTGCGGCGAAGGAAAGGAAGGTGAGGGCGAGGAGCCAGATGGAGGTGGAG CTCTTGGCGATGCTGCTGTGCTCGTTCGAGCATGCCTGGCCGTGGATCCCGCTGCGAGACCGACGTTTGAAAAAA TCATGCAATCCCGATTCTTGGCTGGAAAGGGCGGATGGGGAATGGAGAAGCCAATGTGATTTTCCCGACATAGTA GACGTCCATTTTCGCACTAGATGAAGGCGATTGGGAGTCAGTCTTGTGACAGTAATGTTATCGATAGTGATTGTG ACCGGCGAAATGTACCATCCCGTGTCTGTATTCTGTGATTGAAACTAGCCTATGTAGACTTAGGAAGATGCCAAT GTTGTAGAGTAGATATTACAGTGTACCTCTAACTCATTGTCCTATTTATTGC |
| Length | 2227 |