CopciAB_416566
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_416566 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | rio2 | Synonyms | 416566 |
| Uniprot id | Functional description | Rio2, N-terminal | |
| Location | scaffold_1:2582661..2584597 | Strand | + |
| Gene length (nt) | 1937 | Transcript length (nt) | 1517 |
| CDS length (nt) | 1419 | Protein length (aa) | 472 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YNL207W_RIO2 |
| Aspergillus nidulans | AN0124_rio2 |
| Schizosaccharomyces pombe | rio2 |
| Neurospora crassa | rbg-40 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Schizophyllum commune H4-8 | Schco3_2611113 | 68.6 | 7.211E-215 | 666 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28801 | 67 | 4.02E-213 | 661 |
| Agrocybe aegerita | Agrae_CAA7266509 | 68.2 | 8.651E-212 | 657 |
| Flammulina velutipes | Flave_chr08AA00873 | 66.6 | 7.642E-212 | 657 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_117292 | 66.8 | 6.971E-211 | 654 |
| Lentinula edodes B17 | Lened_B_1_1_3441 | 68.1 | 7.153E-211 | 654 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_2477 | 68.1 | 8.971E-211 | 654 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_182374 | 67.1 | 3.293E-210 | 652 |
| Auricularia subglabra | Aurde3_1_1143383 | 67.7 | 4.228E-207 | 644 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_113901 | 61.8 | 7.548E-203 | 631 |
| Grifola frondosa | Grifr_OBZ79160 | 58.1 | 2.588E-197 | 615 |
| Lentinula edodes NBRC 111202 | Lenedo1_1192762 | 82.2 | 1.125E-180 | 567 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1527618 | 83.2 | 6.07E-179 | 562 |
| Pleurotus ostreatus PC9 | PleosPC9_1_85680 | 82.9 | 5.503E-178 | 559 |
| Pleurotus ostreatus PC15 | PleosPC15_2_23655 | 82.8 | 2.762E-177 | 557 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | rio2 |
|---|---|
| Protein id | CopciAB_416566.T0 |
| Description | Rio2, N-terminal |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd05144 | RIO2_C | IPR030484 | 94 | 269 |
| Pfam | PF09202 | Rio2, N-terminal | IPR015285 | 8 | 90 |
| Pfam | PF01163 | RIO1 family | - | 107 | 279 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR018935 | RIO kinase, conserved site |
| IPR000687 | RIO kinase |
| IPR036388 | Winged helix-like DNA-binding domain superfamily |
| IPR015285 | RIO2 kinase winged helix domain, N-terminal |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR030484 | Serine/threonine-protein kinase Rio2 |
| IPR036390 | Winged helix DNA-binding domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004674 | protein serine/threonine kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004672 | protein kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K07179 |
EggNOG
| COG category | Description |
|---|---|
| T | Rio2, N-terminal |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| winged helix-turn-helix |
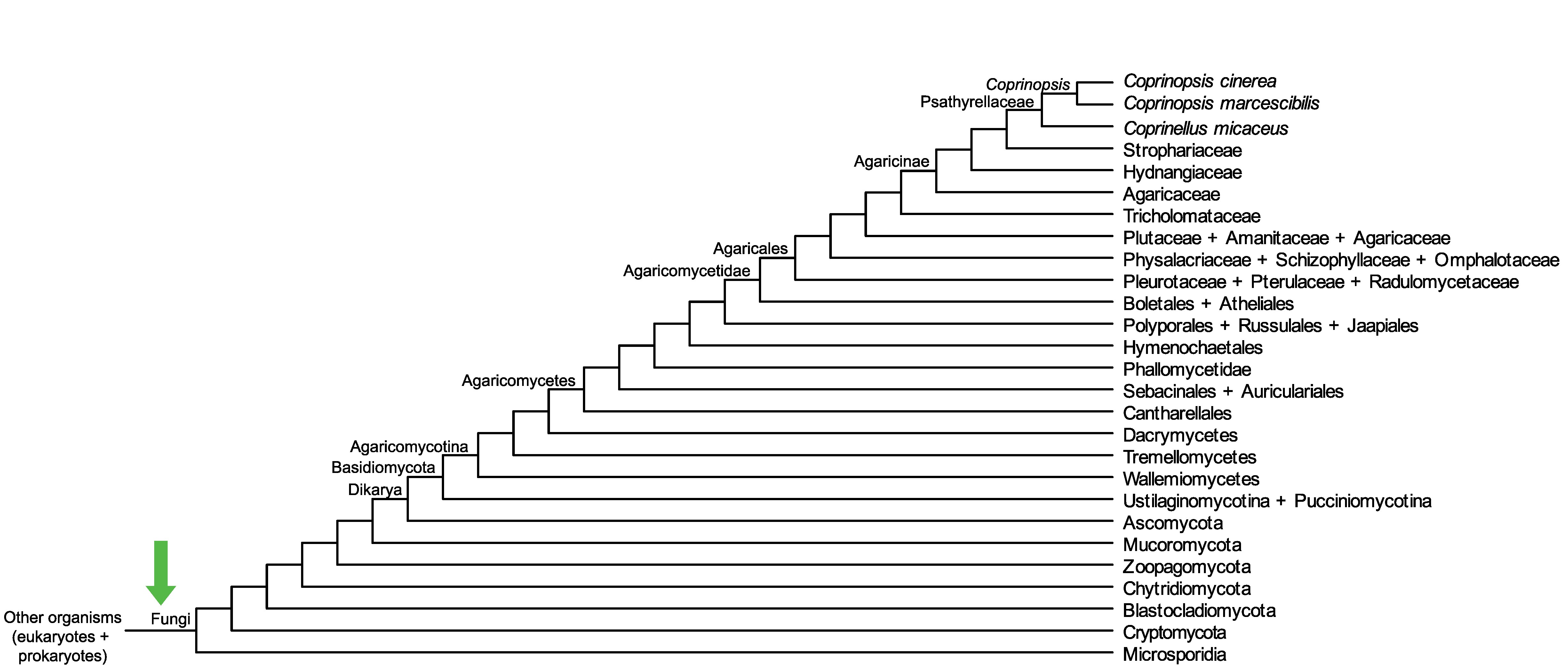
Conservation of CopciAB_416566 across fungi.
Arrow shows the origin of gene family containing CopciAB_416566.
Protein
| Sequence id | CopciAB_416566.T0 |
|---|---|
| Sequence |
>CopciAB_416566.T0 MKLDATDLRYVTSEEFRVLTAVEMGSKNHEIVPTHLIVQISGLRGGGVNKLIGSLAKRNLISKVQNAKYEGYRLT YGGYDYLAMRALSKRDSMTSVGNQIGVGKESDIYVVANAEGDEMVLKLHRLGRISFRSIKTKRDYMGKRKSASWM YLSRLSAQKEYAFMKVLYEHGFPIPKPIDQARHCVLMEFINAYPLRQISEVPSPGKLYSTLMDLIVRFAQAGLIH GDFNEFNILIREETGEPVVIDFPQMVSTSHENAEWYFNRDVECIRTFFRRRFRYESNLYPRFKRTMAEGSADGND FRLDVIVEASGFSKNDMKVLEEYMEAADLEEEGTGDEEDESEESEEEEEEEEEEEEEGEESEEGGLSMPAQRTYD GPSEGESRQKTSESTQQEDGTPDISVLNISRESSRSASPEPDTVKDDIKARAAAEVSKRQNQQRRKYHSKKSVRN AGRPQGSKGKQDTRVKLDSFWA |
| Length | 472 |
Coding
| Sequence id | CopciAB_416566.T0 |
|---|---|
| Sequence |
>CopciAB_416566.T0 ATGAAGCTGGATGCAACAGATCTGAGATATGTCACCTCGGAGGAGTTCCGAGTCCTCACCGCTGTCGAAATGGGC TCGAAAAACCATGAAATTGTCCCCACACACTTGATAGTCCAGATATCGGGGCTTCGAGGAGGAGGAGTAAACAAG CTCATTGGTTCTCTGGCGAAGAGAAACCTTATCTCCAAGGTTCAGAACGCAAAATACGAAGGATATCGCCTTACA TATGGAGGGTACGATTACCTGGCTATGCGCGCATTGTCCAAGAGGGACAGCATGACGTCTGTTGGGAATCAAATT GGCGTTGGAAAGGAATCCGACATCTATGTCGTTGCCAATGCTGAAGGCGACGAGATGGTCCTAAAACTCCACAGA CTAGGACGAATTTCTTTCCGGAGTATCAAAACAAAGCGAGACTATATGGGTAAACGAAAATCTGCATCCTGGATG TACCTCTCGCGTCTCTCCGCGCAGAAAGAATATGCCTTCATGAAAGTGCTTTATGAACATGGTTTCCCCATACCA AAGCCCATCGACCAAGCCAGACATTGCGTACTGATGGAATTCATAAATGCCTACCCGTTGCGACAGATATCAGAA GTGCCGTCTCCAGGAAAGCTCTATTCCACGTTGATGGACCTCATCGTCCGCTTTGCACAAGCAGGCCTTATCCAC GGCGATTTCAACGAATTCAACATCCTCATTCGGGAGGAAACAGGCGAACCCGTGGTCATTGATTTCCCTCAAATG GTTAGCACGTCTCATGAGAACGCAGAATGGTATTTCAACCGCGACGTAGAATGCATCCGCACATTCTTCCGACGA CGGTTCCGCTACGAAAGCAATCTTTATCCTCGTTTCAAAAGGACCATGGCTGAAGGGTCTGCAGATGGCAATGAT TTCCGTTTAGATGTCATCGTGGAGGCCAGCGGATTCAGCAAGAATGACATGAAGGTGTTGGAGGAGTACATGGAG GCTGCGGATCTGGAGGAAGAGGGAACAGGAGATGAAGAGGATGAGAGCGAGGAGAGCGAGGAGGAGGAAGAGGAG GAAGAGGAGGAAGAGGAGGAAGGAGAGGAAAGCGAGGAAGGCGGCCTTAGTATGCCTGCGCAAAGGACATACGAT GGTCCTTCAGAAGGGGAATCACGGCAAAAGACCTCCGAGAGCACCCAGCAGGAAGATGGAACGCCTGACATTTCC GTCCTGAACATATCAAGGGAGTCTTCAAGGTCAGCTTCTCCAGAACCCGACACCGTCAAGGATGATATCAAGGCG AGGGCAGCTGCCGAAGTCTCGAAGCGGCAGAATCAACAAAGACGCAAGTATCACTCCAAAAAGTCTGTTCGAAAT GCGGGCCGTCCACAGGGTAGCAAGGGGAAACAGGATACACGAGTCAAACTGGATAGTTTCTGGGCGTAA |
| Length | 1419 |
Transcript
| Sequence id | CopciAB_416566.T0 |
|---|---|
| Sequence |
>CopciAB_416566.T0 ATCTCTTACTGCACAGTCAAAAACACTGCTCGCTATGAAGCTGGATGCAACAGATCTGAGATATGTCACCTCGGA GGAGTTCCGAGTCCTCACCGCTGTCGAAATGGGCTCGAAAAACCATGAAATTGTCCCCACACACTTGATAGTCCA GATATCGGGGCTTCGAGGAGGAGGAGTAAACAAGCTCATTGGTTCTCTGGCGAAGAGAAACCTTATCTCCAAGGT TCAGAACGCAAAATACGAAGGATATCGCCTTACATATGGAGGGTACGATTACCTGGCTATGCGCGCATTGTCCAA GAGGGACAGCATGACGTCTGTTGGGAATCAAATTGGCGTTGGAAAGGAATCCGACATCTATGTCGTTGCCAATGC TGAAGGCGACGAGATGGTCCTAAAACTCCACAGACTAGGACGAATTTCTTTCCGGAGTATCAAAACAAAGCGAGA CTATATGGGTAAACGAAAATCTGCATCCTGGATGTACCTCTCGCGTCTCTCCGCGCAGAAAGAATATGCCTTCAT GAAAGTGCTTTATGAACATGGTTTCCCCATACCAAAGCCCATCGACCAAGCCAGACATTGCGTACTGATGGAATT CATAAATGCCTACCCGTTGCGACAGATATCAGAAGTGCCGTCTCCAGGAAAGCTCTATTCCACGTTGATGGACCT CATCGTCCGCTTTGCACAAGCAGGCCTTATCCACGGCGATTTCAACGAATTCAACATCCTCATTCGGGAGGAAAC AGGCGAACCCGTGGTCATTGATTTCCCTCAAATGGTTAGCACGTCTCATGAGAACGCAGAATGGTATTTCAACCG CGACGTAGAATGCATCCGCACATTCTTCCGACGACGGTTCCGCTACGAAAGCAATCTTTATCCTCGTTTCAAAAG GACCATGGCTGAAGGGTCTGCAGATGGCAATGATTTCCGTTTAGATGTCATCGTGGAGGCCAGCGGATTCAGCAA GAATGACATGAAGGTGTTGGAGGAGTACATGGAGGCTGCGGATCTGGAGGAAGAGGGAACAGGAGATGAAGAGGA TGAGAGCGAGGAGAGCGAGGAGGAGGAAGAGGAGGAAGAGGAGGAAGAGGAGGAAGGAGAGGAAAGCGAGGAAGG CGGCCTTAGTATGCCTGCGCAAAGGACATACGATGGTCCTTCAGAAGGGGAATCACGGCAAAAGACCTCCGAGAG CACCCAGCAGGAAGATGGAACGCCTGACATTTCCGTCCTGAACATATCAAGGGAGTCTTCAAGGTCAGCTTCTCC AGAACCCGACACCGTCAAGGATGATATCAAGGCGAGGGCAGCTGCCGAAGTCTCGAAGCGGCAGAATCAACAAAG ACGCAAGTATCACTCCAAAAAGTCTGTTCGAAATGCGGGCCGTCCACAGGGTAGCAAGGGGAAACAGGATACACG AGTCAAACTGGATAGTTTCTGGGCGTAAGTAACGGATGACGAACCCCTAGTTAAATTGCCGCCCTGCAATGTAAT TTAGTTCCCACGCCTTC |
| Length | 1517 |
Gene
| Sequence id | CopciAB_416566.T0 |
|---|---|
| Sequence |
>CopciAB_416566.T0 ATCTCTTACTGCACAGTCAAAAACACTGCTCGCTATGAAGCTGGATGCAACAGATCTGAGATATGTCACCTCGGA GGAGTTCCGAGTCCTCACCGCTGTACGCTCGCTCATGATCTTCATATGTAAAACTGCAATTTACCCCATATCCTA GGTCGAAATGGGCTCGAAAAACCATGAAATTGTCCCCACACACTTGATAGTCCAGATATCGGGGCTTCGAGGAGG AGGAGTAAACAAGCTCATTGGTTCTCTGGCGAAGAGAAACCTTATCTCCAAGGTTCAGAACGCAAAATGTGAATA ATTTCCATGTTATCCTGCTAAACGGACCAATAATGACATTGGCTGACTCCAGACGAAGGATATCGCCTTACATAT GGAGGGTACGATTACCTGGCTATGCGCGCATTGTCCAAGAGGGACAGCATGACGTCTGTTGGGAATCAAATTGGC GTTGGAAAGGAATCCGGTGTGTAGTCAATCGTTGACGTCGAGGCCAAGTTCTAATCCCCTTGCTCAGACATCTAT GTCGTTGCCAATGCTGAAGGCGACGAGATGGTCCTAAAACTCCACAGGTGAGTGTGGTTCTCTGTTCGCGTGTCT TTGTGACTGACCGTCTACACAGACTAGGACGAATTTCTTTCCGGAGTATCAAAACAAAGCGAGACTATATGGGTA AACGAAAATCTGCATCCTGGATGTACCTCTCGCGTCTCTCCGCGCAGAAAGAATATGCCTTCATGAAAGTATATA CGTCTCTCTTCTCCTCCGTCCTTGAGAATTCACCCTCGATTAGGTGCTTTATGAACATGGTTTCCCCATACCAAA GCCCATCGACCAAGCCAGACATTGCGTACTGATGGAATTCATAAATGCCTACCCGTTGTAAGATTATTTTCCCTA TGAGATCTCATGCCCCAATCTCACATGACAATAGGCGACAGATATCAGAAGTGCCGTCTCCAGGAAAGCTCTATT CCACGTTGATGGACCTCATCGTCCGCTTTGCACAAGCAGGCCTTATCCACGGCGATTTCAACGAATTCAACATCC TCATTCGGGAGGAAACAGGCGAACCCGTGGTCATTGATTTCCCTCAAATGGTTAGCACGTCTCATGAGAACGCAG AATGGTACGAGGCACTGTCCCCCTTCAACAGAGGATCTTTACCTTCGTCCCTCCAGGTATTTCAACCGCGACGTA GAATGCATCCGCACATTCTTCCGACGACGGTTCCGCTACGAAAGCAATCTTTATCCTCGTTTCAAAAGGACCATG GCTGAAGGGTCTGCAGATGGCAATGATTTCCGTTTAGATGTCATCGTGGAGGCCAGCGGATTCAGCAAGAATGAC ATGAAGGTGTTGGAGGAGGTAGTGTCGACATTTTTTCTATCCCCGAAGTGACGCTGAACTCTCCTTACAGTACAT GGAGGCTGCGGATCTGGAGGAAGAGGGAACAGGAGATGAAGAGGATGAGAGCGAGGAGAGCGAGGAGGAGGAAGA GGAGGAAGAGGAGGAAGAGGAGGAAGGAGAGGAAAGCGAGGAAGGCGGCCTTAGTATGCCTGCGCAAAGGACATA CGATGGTCCTTCAGAAGGGGAATCACGGCAAAAGACCTCCGAGAGCACCCAGCAGGAAGATGGAACGCCTGACAT TTCCGTCCTGAACATATCAAGGGAGTCTTCAAGGTCAGCTTCTCCAGAACCCGACACCGTCAAGGATGATATCAA GGCGAGGGCAGCTGCCGAAGTCTCGAAGCGGCAGAATCAACAAAGACGCAAGTATCACTCCAAAAAGTCTGTTCG AAATGCGGGCCGTCCACAGGGTAGCAAGGGGAAACAGGATACACGAGTCAAACTGGATAGTTTCTGGGCGTAAGT AACGGATGACGAACCCCTAGTTAAATTGCCGCCCTGCAATGTAATTTAGTTCCCACGCCTTC |
| Length | 1937 |