CopciAB_417694
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_417694 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | ppk32 | Synonyms | 417694 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_2:2124801..2128182 | Strand | - |
| Gene length (nt) | 3382 | Transcript length (nt) | 2828 |
| CDS length (nt) | 2625 | Protein length (aa) | 874 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN4322_scy1 |
| Schizosaccharomyces pombe | ppk32 |
| Neurospora crassa | stk-32 |
| Saccharomyces cerevisiae | YGL083W_SCY1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_199815 | 66.8 | 0 | 1135 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_53378 | 66.7 | 0 | 1135 |
| Agrocybe aegerita | Agrae_CAA7267966 | 65.9 | 0 | 1116 |
| Pleurotus ostreatus PC9 | PleosPC9_1_81775 | 65 | 0 | 1113 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21346 | 66.7 | 0 | 1112 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1073929 | 66.3 | 0 | 1107 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1451782 | 64.7 | 0 | 1102 |
| Flammulina velutipes | Flave_chr09AA00122 | 65.2 | 0 | 1085 |
| Lentinula edodes NBRC 111202 | Lenedo1_1092730 | 64.4 | 0 | 1082 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_89312 | 64.6 | 0 | 1060 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_2688 | 61.7 | 0 | 1032 |
| Grifola frondosa | Grifr_OBZ75157 | 66.5 | 1.482E-291 | 1002 |
| Auricularia subglabra | Aurde3_1_1325094 | 58.8 | 7.46E-304 | 957 |
| Schizophyllum commune H4-8 | Schco3_1087164 | 64.6 | 5.669E-304 | 954 |
| Lentinula edodes B17 | Lened_B_1_1_14875 | 74.7 | 1.519E-248 | 784 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | ppk32 |
|---|---|
| Protein id | CopciAB_417694.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14011 | PK_SCY1_like | - | 62 | 332 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 99 | 332 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR011989 | Armadillo-like helical |
| IPR016024 | Armadillo-type fold |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K17541 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
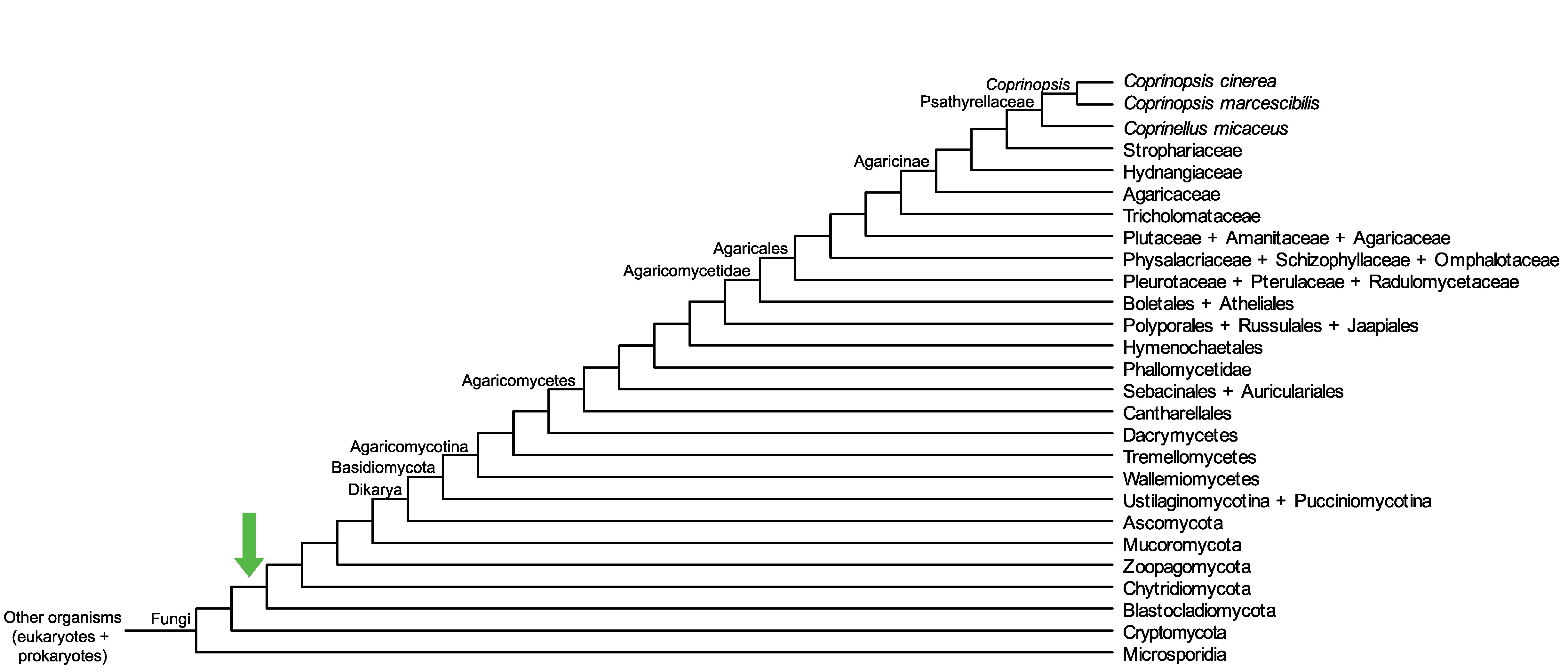
Conservation of CopciAB_417694 across fungi.
Arrow shows the origin of gene family containing CopciAB_417694.
Protein
| Sequence id | CopciAB_417694.T0 |
|---|---|
| Sequence |
>CopciAB_417694.T0 MAHVLAAASSFFGRTNISQSYNIGNPSQTSRPSTPGTTSGGSSGGGGANIAPLPFTPTFYIGLWKVQPATHKVTG KRVSVWSFDKRGPEMERLGVAGKDRVLEVLKNEASALSRLRHPSILEMVEPLEETRIELVFATEPVLSSLELSIP GSGRHASLVELDEIEIQKGILQLAKGLSFLHSQAQLIHSNICPESIIINSSGDWKISGLGVTIPLLTNGNPTRWE FPTFDGRVPAYIQRSFDYMAPEYALDEQLLTSSDMYSLGCVVYAVHSKGNPPFKNHGSLSNLRENAGKPLPGMER WEQDLQSLVRSLVTRHATSRPTPQTLSSHPFFSSLPISTLNFLDRSNFTAKTREEKISFMKGLTGVLNRFSEGLQ TRKILPSLLEEMKDTHLLPYILPNVFAISQALSPTQFASMVLPSLKPLFAIKEPPQNMLTLLDNLGMLQEKTEKN VFKDHVLPLVYNALESEHAIVQERALKAVPDLCETVDYAEVQGVLFPRVAVVFSKTRILSVKVATLVTFLAMVKT LDQSSLTQKLVPLLSKIRTKEPSVMMATLDVQEAMGFKVDREAVATLVLPQLWAMSMGPLLNVSQFQRFMSVIKK LGERVEREHDQFLRDSQRLEDRSATATMGNGGVPLNTTTADFETLVSKGSAMANGGLASSAVTATPSAAPAPAAS WEDDVWGSLLNGDQTAKHQTPTVSSSFSSPSLQTPPVITPSLSPPQQPSRPNFQPRPSKLGTSQPASRLSSSSFG STASTSSKPNYNITLSTAPITTATPSASSLPPAPGFSSPAASSQPSYAAPNYNINLGSSLTTQAPSIQPSVFSTP LVASTPAAPISMGIGGNILTPSKPAQPAWGAGGQKQLSKADWGDFDPLA |
| Length | 874 |
Coding
| Sequence id | CopciAB_417694.T0 |
|---|---|
| Sequence |
>CopciAB_417694.T0 ATGGCACACGTATTAGCAGCAGCAAGCTCGTTCTTCGGGCGCACAAACATTTCGCAGTCGTATAACATCGGAAAT CCGTCACAGACCAGTCGACCATCGACGCCGGGAACGACCTCTGGTGGTAGCAGTGGCGGAGGAGGTGCCAACATC GCCCCGCTGCCCTTCACACCTACTTTTTATATTGGTTTATGGAAAGTACAGCCTGCAACACACAAGGTGACGGGG AAACGCGTCTCCGTGTGGTCGTTTGACAAACGGGGTCCAGAGATGGAGAGGTTGGGGGTTGCCGGGAAGGACAGA GTACTCGAGGTGTTGAAGAATGAGGCGTCCGCACTAAGTCGACTACGGCATCCTTCAATTCTGGAAATGGTGGAA CCGTTGGAAGAAACACGAATAGAGCTTGTATTTGCAACGGAGCCGGTTCTATCGTCCCTGGAATTGTCGATACCA GGTTCGGGCAGACATGCTTCATTGGTGGAACTGGACGAGATCGAGATCCAGAAGGGCATCTTGCAATTGGCAAAG GGACTTTCGTTCTTGCACAGCCAAGCGCAATTAATTCATTCGAACATCTGCCCGGAGAGCATTATCATCAATAGC TCCGGCGATTGGAAGATATCTGGACTAGGAGTAACCATCCCTCTTCTTACTAACGGCAACCCAACTCGTTGGGAA TTTCCGACATTCGACGGCCGTGTGCCAGCATATATCCAGCGGTCATTCGATTATATGGCGCCGGAATATGCTCTT GACGAACAGCTTTTAACAAGTTCGGACATGTACTCTTTAGGCTGCGTGGTCTACGCCGTCCATAGCAAGGGTAAT CCTCCCTTCAAGAACCATGGAAGCCTTAGCAACCTCCGAGAAAATGCCGGGAAACCTTTGCCCGGAATGGAGAGA TGGGAACAGGATTTGCAATCGCTCGTTCGTTCGCTGGTCACCCGTCATGCTACATCACGGCCGACGCCTCAAACA TTATCTTCCCACCCCTTCTTCTCATCACTTCCAATTTCCACCCTGAACTTCCTGGATCGGTCGAACTTCACAGCT AAAACCCGCGAAGAGAAGATATCCTTCATGAAAGGACTGACAGGGGTGCTAAATCGATTCTCGGAAGGTTTGCAA ACCAGGAAGATTCTGCCATCTTTACTCGAGGAGATGAAAGATACCCATTTACTACCATACATTCTTCCTAACGTC TTTGCTATCTCCCAAGCACTGTCTCCGACCCAGTTCGCGTCGATGGTCCTTCCAAGTCTCAAACCTCTTTTCGCA ATCAAGGAACCTCCCCAGAACATGCTGACCCTACTCGACAATCTTGGAATGTTGCAAGAGAAGACGGAGAAGAAT GTATTCAAGGACCACGTCCTACCTCTGGTGTATAATGCCCTTGAGTCTGAACACGCAATCGTTCAGGAGCGGGCG TTGAAAGCAGTTCCCGACCTATGTGAAACGGTTGACTATGCTGAGGTCCAGGGTGTTCTGTTCCCTCGCGTCGCT GTTGTATTTTCCAAGACTAGGATTTTGTCAGTGAAGGTCGCAACATTGGTCACATTTTTAGCAATGGTGAAGACA TTGGATCAGTCAAGTCTGACGCAGAAACTGGTCCCGTTACTTTCGAAAATCAGAACAAAGGAGCCGTCAGTGATG ATGGCCACCTTGGATGTGCAAGAAGCCATGGGTTTCAAGGTGGATCGAGAAGCCGTCGCCACTCTCGTTCTCCCG CAACTATGGGCGATGTCAATGGGTCCTCTACTTAATGTCTCGCAATTCCAGCGTTTCATGTCTGTGATCAAGAAG CTTGGAGAACGGGTTGAACGGGAACATGATCAATTCCTGCGTGATTCCCAACGACTGGAAGATCGGTCTGCTACC GCTACTATGGGGAACGGTGGAGTTCCTCTGAACACAACCACAGCCGATTTCGAAACCCTAGTAAGCAAGGGCAGT GCGATGGCCAATGGAGGTTTGGCTAGTTCTGCAGTCACCGCCACCCCTAGCGCTGCCCCTGCTCCTGCTGCGTCA TGGGAAGACGACGTCTGGGGTAGCTTGCTGAACGGAGACCAGACAGCGAAGCATCAGACTCCTACTGTTAGCAGC AGCTTTAGTTCCCCTTCCCTCCAGACGCCGCCGGTGATCACACCATCCCTCTCGCCGCCACAGCAACCCAGCCGA CCCAATTTTCAGCCGCGGCCTTCGAAGCTTGGGACATCACAACCGGCATCCAGGTTATCGTCATCTTCGTTCGGG TCCACTGCTTCCACCTCGTCCAAGCCGAATTACAACATCACTCTGTCCACTGCCCCTATCACTACCGCTACACCA TCTGCCTCTTCTCTCCCACCCGCTCCTGGCTTCAGTTCCCCGGCAGCTTCTTCACAGCCAAGCTATGCCGCACCC AATTACAATATCAACCTTGGATCGTCATTAACCACCCAAGCGCCTTCCATACAACCTTCAGTATTCTCCACACCT CTGGTTGCGAGCACTCCTGCCGCTCCAATAAGTATGGGAATAGGTGGGAACATCCTCACGCCATCCAAGCCCGCA CAACCGGCGTGGGGCGCAGGCGGACAGAAACAGCTGTCGAAAGCAGACTGGGGAGACTTTGACCCGCTAGCGTAG |
| Length | 2625 |
Transcript
| Sequence id | CopciAB_417694.T0 |
|---|---|
| Sequence |
>CopciAB_417694.T0 ACGACTGTCAATACCGACCACCAACACCGCTAATCCTTCGGATTCTTCCAAAGGTCAACGACGACCACAACGCCT GACGATGGCACACGTATTAGCAGCAGCAAGCTCGTTCTTCGGGCGCACAAACATTTCGCAGTCGTATAACATCGG AAATCCGTCACAGACCAGTCGACCATCGACGCCGGGAACGACCTCTGGTGGTAGCAGTGGCGGAGGAGGTGCCAA CATCGCCCCGCTGCCCTTCACACCTACTTTTTATATTGGTTTATGGAAAGTACAGCCTGCAACACACAAGGTGAC GGGGAAACGCGTCTCCGTGTGGTCGTTTGACAAACGGGGTCCAGAGATGGAGAGGTTGGGGGTTGCCGGGAAGGA CAGAGTACTCGAGGTGTTGAAGAATGAGGCGTCCGCACTAAGTCGACTACGGCATCCTTCAATTCTGGAAATGGT GGAACCGTTGGAAGAAACACGAATAGAGCTTGTATTTGCAACGGAGCCGGTTCTATCGTCCCTGGAATTGTCGAT ACCAGGTTCGGGCAGACATGCTTCATTGGTGGAACTGGACGAGATCGAGATCCAGAAGGGCATCTTGCAATTGGC AAAGGGACTTTCGTTCTTGCACAGCCAAGCGCAATTAATTCATTCGAACATCTGCCCGGAGAGCATTATCATCAA TAGCTCCGGCGATTGGAAGATATCTGGACTAGGAGTAACCATCCCTCTTCTTACTAACGGCAACCCAACTCGTTG GGAATTTCCGACATTCGACGGCCGTGTGCCAGCATATATCCAGCGGTCATTCGATTATATGGCGCCGGAATATGC TCTTGACGAACAGCTTTTAACAAGTTCGGACATGTACTCTTTAGGCTGCGTGGTCTACGCCGTCCATAGCAAGGG TAATCCTCCCTTCAAGAACCATGGAAGCCTTAGCAACCTCCGAGAAAATGCCGGGAAACCTTTGCCCGGAATGGA GAGATGGGAACAGGATTTGCAATCGCTCGTTCGTTCGCTGGTCACCCGTCATGCTACATCACGGCCGACGCCTCA AACATTATCTTCCCACCCCTTCTTCTCATCACTTCCAATTTCCACCCTGAACTTCCTGGATCGGTCGAACTTCAC AGCTAAAACCCGCGAAGAGAAGATATCCTTCATGAAAGGACTGACAGGGGTGCTAAATCGATTCTCGGAAGGTTT GCAAACCAGGAAGATTCTGCCATCTTTACTCGAGGAGATGAAAGATACCCATTTACTACCATACATTCTTCCTAA CGTCTTTGCTATCTCCCAAGCACTGTCTCCGACCCAGTTCGCGTCGATGGTCCTTCCAAGTCTCAAACCTCTTTT CGCAATCAAGGAACCTCCCCAGAACATGCTGACCCTACTCGACAATCTTGGAATGTTGCAAGAGAAGACGGAGAA GAATGTATTCAAGGACCACGTCCTACCTCTGGTGTATAATGCCCTTGAGTCTGAACACGCAATCGTTCAGGAGCG GGCGTTGAAAGCAGTTCCCGACCTATGTGAAACGGTTGACTATGCTGAGGTCCAGGGTGTTCTGTTCCCTCGCGT CGCTGTTGTATTTTCCAAGACTAGGATTTTGTCAGTGAAGGTCGCAACATTGGTCACATTTTTAGCAATGGTGAA GACATTGGATCAGTCAAGTCTGACGCAGAAACTGGTCCCGTTACTTTCGAAAATCAGAACAAAGGAGCCGTCAGT GATGATGGCCACCTTGGATGTGCAAGAAGCCATGGGTTTCAAGGTGGATCGAGAAGCCGTCGCCACTCTCGTTCT CCCGCAACTATGGGCGATGTCAATGGGTCCTCTACTTAATGTCTCGCAATTCCAGCGTTTCATGTCTGTGATCAA GAAGCTTGGAGAACGGGTTGAACGGGAACATGATCAATTCCTGCGTGATTCCCAACGACTGGAAGATCGGTCTGC TACCGCTACTATGGGGAACGGTGGAGTTCCTCTGAACACAACCACAGCCGATTTCGAAACCCTAGTAAGCAAGGG CAGTGCGATGGCCAATGGAGGTTTGGCTAGTTCTGCAGTCACCGCCACCCCTAGCGCTGCCCCTGCTCCTGCTGC GTCATGGGAAGACGACGTCTGGGGTAGCTTGCTGAACGGAGACCAGACAGCGAAGCATCAGACTCCTACTGTTAG CAGCAGCTTTAGTTCCCCTTCCCTCCAGACGCCGCCGGTGATCACACCATCCCTCTCGCCGCCACAGCAACCCAG CCGACCCAATTTTCAGCCGCGGCCTTCGAAGCTTGGGACATCACAACCGGCATCCAGGTTATCGTCATCTTCGTT CGGGTCCACTGCTTCCACCTCGTCCAAGCCGAATTACAACATCACTCTGTCCACTGCCCCTATCACTACCGCTAC ACCATCTGCCTCTTCTCTCCCACCCGCTCCTGGCTTCAGTTCCCCGGCAGCTTCTTCACAGCCAAGCTATGCCGC ACCCAATTACAATATCAACCTTGGATCGTCATTAACCACCCAAGCGCCTTCCATACAACCTTCAGTATTCTCCAC ACCTCTGGTTGCGAGCACTCCTGCCGCTCCAATAAGTATGGGAATAGGTGGGAACATCCTCACGCCATCCAAGCC CGCACAACCGGCGTGGGGCGCAGGCGGACAGAAACAGCTGTCGAAAGCAGACTGGGGAGACTTTGACCCGCTAGC GTAGTCCACTCTGTAACGATTTACTACATGCCGCATTCCTTCACGGAATGCCTGGTCACTGCTTATTTATGAGAT CAATCACTGGGGGTTTCATACCACGACTATGGTGTAATCGCCATGCTTATACA |
| Length | 2828 |
Gene
| Sequence id | CopciAB_417694.T0 |
|---|---|
| Sequence |
>CopciAB_417694.T0 ACGACTGTCAATACCGACCACCAACACCGCTAATCCTTCGGATTCTTCCAAAGGTCAACGACGACCACAACGCCT GACGATGGCACACGTATTAGCAGCAGCAAGCTCGTTCTTCGGGCGCACAAACATTTCGCAGTCGTATAACATCGG AAATCCGTCACAGACCAGTCGACCATCGACGCCGGGAACGACCTCTGGTGGTAGCAGTGGCGGAGGAGGTGCCAA CATCGCCCCGCTGCCCTTCACACCTACTTTTTATATTGGTTTATGGAAAGTACAGCCTGCAACACACAAGGTGAC GGGGAAACGCGTCTCCGTGTGGTCGTTTGACAAACGGGGTCCAGAGATGGAGAGGTTGGGGGTTGCCGGGAAGGA CAGAGTACTCGAGGTGTTGAAGAATGAGGTGCGATGATCTTCAATCCCCTTTAATCAGACTTCCGGTTTAACGCT CGAGTTCAGGCGTCCGCACTAAGTCGACTACGGCATCCTTCAATTCTGGGTACGTGGAGTTGACAGAATTCGACC GAATCAGCGCTTATTGTGACTTTGGCCCACAGAAATGGTGGAACCGTTGGAAGAAACACGAATAGAGCTTGTATT TGCAACGGAGCCGGTTCTATCGTCCCTGGAATTGTCGATACCAGGTTCGGGCAGACATGCTTCATTGGTGGAACT GGACGAGATCGAGGTGGGCGCGTGATCCTCGAGTGGGTGCTAGTGTCTAATTGTGCGGTTAGATCCAGAAGGGCA TCTTGCAATTGGCAAAGGGACTTTCGTTCTTGCACAGCCAAGCGCAATTAATTCATTCGAACATCTGCCCGGAGA GCATTATCATCAATAGCTCCGTACGTCCAGCACTTCATCTGCAGCTGTACCGCCTTGTGCCAACCAGTACCATAG GGCGATTGGAAGATATCTGGACTAGGAGTAACCATCCCTCTTCTTACTAACGGCAACCCAACTCGTTGGGAATTT CCGACATTCGACGGCCGTGTGCCAGCATATATCCAGCGGTCATTCGATTATATGGGTACGCAATGTATCCCTTCC ATCCCACCACACCCAGGACACTGACAGGACAAGATAGCGCCGGAATATGCTCTTGACGAACAGCTTTTAACAAGT TCGGACATGTACTCTTTAGGCTGCGTGGTCTACGCCGTCCATAGCAAGGGTAATCCTCCCTTCAAGAACCATGGA AGCCTTAGCAACCTCCGAGAAAATGCCGGGAAACCTTTGCCCGGAATGGAGAGATGGGAACAGGATTTGCAATGT GCGCACCATAATTCTATCCAAACTGTCAAAGTCCCATTGATTTGCCGTGCACCTAGCGCTCGTTCGTTCGCTGGT CACCCGTCATGCTACATCACGGCCGACGCCTCAAACATTATCTTCCCACCCCTTCTTCTCATCACTTCCAATTTC CACCCTGAACTTCCTGGATCGGTCGAACTTCACAGCTAAAACCCGCGAAGAGAAGATATCCTTCATGAAAGGACT GACAGGGGTGCTAAATCGATTCTCGGAAGGTTTGCAAACCAGGAAGATTCTGCCATCTTTACTCGAGGAGGTGAG TCTCATTATCTTGTCTACTCCATTGCTCGCTTACCCGCGGTGCTTGTTAGATGAAAGATACCCATTTACTACCAT ACATTCTTCCTAACGTCTTTGCTATCTCCCAAGCACTGTCTCCGACCCAGTTCGCGTCGATGGTCCTTCCAAGTC TCAAACCTCTTTTCGCAATCAAGGAACCTCCCCAGAACATGCTGACCCTACTCGACAATCTTGGAATGTTGCAAG AGAAGACGGAGAAGAATGTATTCAAGGACCGTGAGTCCCGCCGTTCCTGTTTAGCTTTGCGGGGTCTCAAGGTTT CTCCATGTTAAGACGTCCTACCTCTGGTGTATAATGCCCTTGAGTCTGAACACGCAATCGTTCAGGAGCGGGCGT TGAAAGCAGTTCCCGACCTATGTGAAACGGTTGACTATGCTGAGGTCCAGGGTGTTCTGTTCCCTCGCGTCGCTG TATGTTCCAAGCTTCCTGATGATGCTTAATATTGTTTCTTAACTGTCATGATCTGCAGGTTGTATTTTCCAAGAC TAGGATTTTGTCAGTGAAGGTCGCAACATTGGTCACATTTTTAGCAATGGTGAAGACATTGGATCAGTCAAGTCT GACGCAGAAACTGGTCCCGTTACTTTCGAAAATCAGAACAAAGGAGCCGTCAGTGATGGTAAGTAGAATCCGTTC ATCGTTGCATGGGTTGACGACGGTCTGTTACAGATGGCCACCTTGGATGTGCAAGAAGCCATGGGTTTCAAGGTG GATCGAGAAGCCGTCGCCACTCTCGTTCTCCCGCAACTATGGGCGATGTCAATGGGTCCTCTACTTAATGTCTCG CAATTCCAGCGTTTCATGTCTGTGATCAAGAAGCTTGGAGAACGGGTTGAACGGGAACATGATCAATTCCTGCGT GATTCCCAACGACTGGAAGATCGGTCTGCTACCGCTACTATGGGGAACGGTGGAGTTCCTCTGAACACAACCACA GCCGATTTCGAAACCCTAGTAAGCAAGGGCAGTGCGATGGCCAATGGAGGTTTGGCTAGTTCTGCAGTCACCGCC ACCCCTAGCGCTGCCCCTGCTCCTGCTGCGTCATGGGAAGACGACGTCTGGGGTAGCTTGCTGAACGGAGACCAG ACAGCGAAGCATCAGACTCCTACTGTTAGCAGCAGCTTTAGTTCCCCTTCCCTCCAGACGCCGCCGGTGATCACA CCATCCCTCTCGCCGCCACAGCAACCCAGCCGACCCAATTTTCAGCCGCGGCCTTCGAAGCTTGGGACATCACAA CCGGCATCCAGGTTATCGTCATCTTCGTTCGGGTCCACTGCTTCCACCTCGTCCAAGCCGAATTACAACATCACT CTGTCCACTGCCCCTATCACTACCGCTACACCATCTGCCTCTTCTCTCCCACCCGCTCCTGGCTTCAGTTCCCCG GCAGCTTCTTCACAGCCAAGCTATGCCGCACCCAATTACAATATCAACCTTGGATCGTCATTAACCACCCAAGCG CCTTCCATACAACCTTCAGTATTCTCCACACCTCTGGTTGCGAGCACTCCTGCCGCTCCAATAAGTATGGGAATA GGTGGGAACATCCTCACGCCATCCAAGCCCGCACAACCGGCGTGGGGCGCAGGCGGACAGAAACAGCTGTCGAAA GCAGACTGGGGAGACTTTGACCCGCTAGCGTAGTCCACTCTGTAACGATTTACTACATGCCGCATTCCTTCACGG AATGCCTGGTCACTGCTTATTTATGAGATCAATCACTGGGGGTTTCATACCACGACTATGGTGTAATCGCCATGC TTATACA |
| Length | 3382 |