CopciAB_420995
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_420995 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 420995 |
| Uniprot id | Functional description | ||
| Location | scaffold_10:1723509..1726471 | Strand | - |
| Gene length (nt) | 2963 | Transcript length (nt) | 2656 |
| CDS length (nt) | 2337 | Protein length (aa) | 778 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Flammulina velutipes | Flave_chr07AA00019 | 29.5 | 1.67E-59 | 222 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1503299 | 30.4 | 2.675E-55 | 209 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_220 | 25.2 | 2.387E-54 | 206 |
| Lentinula edodes NBRC 111202 | Lenedo1_26445 | 26.4 | 4.056E-52 | 199 |
| Schizophyllum commune H4-8 | Schco3_2486423 | 26 | 9.441E-46 | 179 |
| Pleurotus ostreatus PC9 | PleosPC9_1_128740 | 25.6 | 3.943E-42 | 167 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1108721 | 23.9 | 3.462E-41 | 164 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_420995.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 198 | 618 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR040976 | Fungal-type protein kinase |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
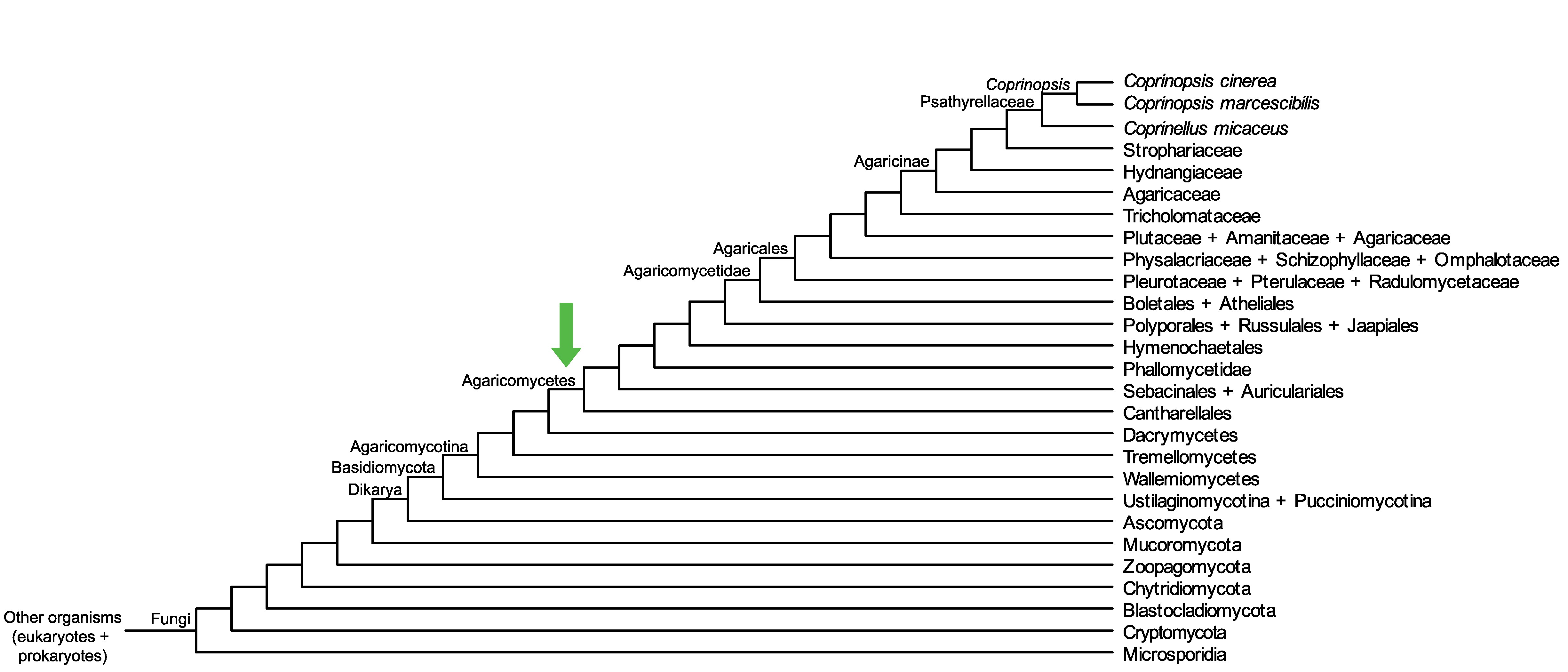
Conservation of CopciAB_420995 across fungi.
Arrow shows the origin of gene family containing CopciAB_420995.
Protein
| Sequence id | CopciAB_420995.T0 |
|---|---|
| Sequence |
>CopciAB_420995.T0 METEQLKASEQDQNYFDSRTAAQKDAEEQRKTHLFNVKDHLLQCDFSTWFETCLSNPDDMKWSEKSALDVKRRLL TLSPPVLTDEGWVPLKKALGETDKKEDTVYASMTGIISKIKHAAAQELKLDSDFEGQITVYSSTPTRPTTNDVHG PNLKPDGRFVLAPPPPQPPTHVLYRRSSRLAKKMEEEARSQRNATASSNVSDISMSTSDSVALFEYKLKEADQRQ NYRQMLGGAAFVLYNDPRRTRVYGVTIEETTTRLWHFDRGAVLFSPTFDCQEKPEYLIRICLYLSYASPIQLGYD PTVKKVESKNGKGHSPDFIYQVNNKYYRTIGQPLSETSAFWTISRGIRTWVVQECDENGAIKPNIAEAVLRDLWL YEDEPDEKAIQTEILSHLSRRNIPVKAMEQLFVHILEDVTLSGKEQRTLAKPVDAFQFPFNPDSGIPRKAPPTES RKRSGGVEQDVFSDYEEVIQPPTPLQHHPRTHRRTLFKERCTPFYGVPNRRVAFEVCKGLVRGLDLLRQAGFLHR NISGSSCVFYYDKDAQAYQAKLIELEYCKRYQDTRFHDPKSVPREFAAVEVMDSQWNLAKTGLDVLQLDELLPPF HRHYYHDLESLFWLLTWYTTTHLPIDRDENQEIVANLDLVSWKTKVFDVLFPRESHLERRRNLWERPNQVYRDLK AAVGWPIDTVTILLSLSKIPSYFDSEYTALYRNPPQDKAVRWPDARFNDSLYKKFINTLDDVAARLGTVGSVSMW GLLNEGRMKNKRPGKEKDETDRIHKKRG |
| Length | 778 |
Coding
| Sequence id | CopciAB_420995.T0 |
|---|---|
| Sequence |
>CopciAB_420995.T0 ATGGAGACAGAACAGTTAAAAGCATCAGAACAGGACCAAAACTACTTCGACAGTCGCACTGCTGCGCAAAAGGAC GCAGAGGAACAGCGCAAAACCCATCTTTTCAATGTGAAAGATCACCTTTTACAGTGTGACTTTTCCACCTGGTTC GAAACCTGCCTTTCGAATCCTGACGATATGAAGTGGAGCGAGAAATCTGCTTTGGATGTCAAGAGGAGACTTCTT ACATTGTCGCCGCCCGTCCTCACAGACGAAGGCTGGGTGCCATTGAAGAAGGCCTTGGGTGAAACCGACAAGAAG GAGGACACCGTTTACGCATCTATGACGGGTATCATTTCGAAAATCAAGCATGCGGCGGCTCAGGAACTTAAGCTT GATTCCGATTTTGAGGGACAAATAACAGTCTATTCCTCCACTCCCACTCGACCAACTACTAACGACGTCCATGGA CCCAACTTGAAGCCTGACGGTCGCTTTGTACTCGCACCACCGCCGCCTCAGCCTCCAACTCATGTTCTCTACCGC CGATCATCGCGGCTAGCGAAAAAGATGGAGGAGGAGGCGCGTAGTCAGAGGAATGCCACTGCAAGCAGCAACGTT TCCGACATATCCATGTCTACGTCTGATTCGGTTGCGCTCTTTGAATACAAGCTCAAGGAGGCAGATCAGCGACAG AACTACCGGCAAATGCTGGGAGGTGCAGCTTTCGTGCTGTACAACGACCCACGTCGCACTCGGGTCTATGGGGTC ACCATTGAAGAAACTACCACGCGGCTGTGGCACTTTGATCGCGGAGCGGTCCTCTTCAGCCCTACGTTTGACTGT CAAGAGAAACCTGAATACCTCATCAGGATCTGCTTGTATCTGTCATACGCGAGCCCGATACAGCTTGGCTACGAC CCCACCGTGAAGAAGGTCGAGAGCAAAAATGGGAAAGGGCATTCACCAGACTTCATCTACCAAGTCAACAACAAG TACTATCGAACCATCGGACAGCCACTGTCTGAAACAAGTGCATTCTGGACAATCTCGAGAGGAATAAGGACTTGG GTCGTCCAGGAGTGCGATGAGAATGGCGCGATCAAGCCAAATATAGCCGAGGCCGTTCTAAGGGACCTATGGCTG TATGAAGATGAGCCTGACGAAAAGGCTATTCAGACTGAGATTCTCAGTCACTTATCCAGACGCAACATTCCAGTC AAGGCGATGGAACAGCTGTTCGTCCATATATTGGAGGATGTCACCCTTTCAGGGAAAGAGCAAAGAACACTGGCC AAACCGGTGGATGCATTCCAATTTCCATTCAACCCTGATTCAGGTATCCCGAGAAAGGCCCCACCGACAGAATCT CGAAAGCGGTCAGGGGGGGTGGAGCAGGATGTATTCTCTGATTATGAAGAGGTCATCCAACCACCGACTCCTCTC CAGCATCATCCACGCACCCATCGTCGCACACTGTTCAAGGAGAGGTGTACGCCATTTTACGGAGTTCCGAATCGC CGGGTTGCATTCGAGGTCTGCAAAGGCCTCGTTCGTGGGCTGGATCTTTTGCGACAAGCAGGATTTTTACATCGG AATATCAGCGGCTCCAGCTGCGTGTTCTACTACGACAAGGATGCCCAAGCCTATCAGGCTAAACTGATCGAGTTG GAGTATTGCAAAAGATATCAAGATACACGTTTCCATGATCCCAAATCGGTTCCACGGGAATTTGCCGCTGTTGAA GTCATGGATTCCCAGTGGAACCTCGCGAAGACCGGTCTGGATGTCCTCCAGTTGGATGAATTACTCCCCCCTTTC CACCGACATTATTACCACGATCTGGAGTCGCTCTTCTGGCTATTAACCTGGTACACAACCACGCATTTGCCTATC GACAGGGATGAAAATCAGGAAATCGTGGCTAATCTTGACCTTGTCAGTTGGAAGACAAAGGTCTTTGATGTTTTG TTCCCGCGGGAATCCCATTTAGAGCGCCGCCGAAACCTATGGGAAAGACCAAACCAGGTTTACCGTGACTTGAAG GCTGCCGTCGGATGGCCAATAGACACGGTCACTATCCTCCTCAGCCTCAGCAAGATACCTTCTTACTTTGATTCG GAATACACGGCACTTTATCGGAATCCTCCACAGGATAAAGCAGTGCGGTGGCCCGATGCAAGGTTCAACGATTCA CTATACAAGAAATTTATCAACACTCTGGACGATGTCGCTGCTCGTCTTGGAACAGTTGGTTCTGTTTCAATGTGG GGTTTACTGAATGAGGGGAGGATGAAGAACAAGAGGCCCGGGAAAGAAAAGGACGAGACTGACAGAATCCATAAG AAGCGGGGATGA |
| Length | 2337 |
Transcript
| Sequence id | CopciAB_420995.T0 |
|---|---|
| Sequence |
>CopciAB_420995.T0 ACCCACACAGACCCACGATTCTCCTGCACGTACCCTCGCCTTCCACTCGTTATCCTCGCTCTCTCTCTTGAACTA TATCTACAAGAATGGAGACAGAACAGTTAAAAGCATCAGAACAGGACCAAAACTACTTCGACAGTCGCACTGCTG CGCAAAAGGACGCAGAGGAACAGCGCAAAACCCATCTTTTCAATGTGAAAGATCACCTTTTACAGTGTGACTTTT CCACCTGGTTCGAAACCTGCCTTTCGAATCCTGACGATATGAAGTGGAGCGAGAAATCTGCTTTGGATGTCAAGA GGAGACTTCTTACATTGTCGCCGCCCGTCCTCACAGACGAAGGCTGGGTGCCATTGAAGAAGGCCTTGGGTGAAA CCGACAAGAAGGAGGACACCGTTTACGCATCTATGACGGGTATCATTTCGAAAATCAAGCATGCGGCGGCTCAGG AACTTAAGCTTGATTCCGATTTTGAGGGACAAATAACAGTCTATTCCTCCACTCCCACTCGACCAACTACTAACG ACGTCCATGGACCCAACTTGAAGCCTGACGGTCGCTTTGTACTCGCACCACCGCCGCCTCAGCCTCCAACTCATG TTCTCTACCGCCGATCATCGCGGCTAGCGAAAAAGATGGAGGAGGAGGCGCGTAGTCAGAGGAATGCCACTGCAA GCAGCAACGTTTCCGACATATCCATGTCTACGTCTGATTCGGTTGCGCTCTTTGAATACAAGCTCAAGGAGGCAG ATCAGCGACAGAACTACCGGCAAATGCTGGGAGGTGCAGCTTTCGTGCTGTACAACGACCCACGTCGCACTCGGG TCTATGGGGTCACCATTGAAGAAACTACCACGCGGCTGTGGCACTTTGATCGCGGAGCGGTCCTCTTCAGCCCTA CGTTTGACTGTCAAGAGAAACCTGAATACCTCATCAGGATCTGCTTGTATCTGTCATACGCGAGCCCGATACAGC TTGGCTACGACCCCACCGTGAAGAAGGTCGAGAGCAAAAATGGGAAAGGGCATTCACCAGACTTCATCTACCAAG TCAACAACAAGTACTATCGAACCATCGGACAGCCACTGTCTGAAACAAGTGCATTCTGGACAATCTCGAGAGGAA TAAGGACTTGGGTCGTCCAGGAGTGCGATGAGAATGGCGCGATCAAGCCAAATATAGCCGAGGCCGTTCTAAGGG ACCTATGGCTGTATGAAGATGAGCCTGACGAAAAGGCTATTCAGACTGAGATTCTCAGTCACTTATCCAGACGCA ACATTCCAGTCAAGGCGATGGAACAGCTGTTCGTCCATATATTGGAGGATGTCACCCTTTCAGGGAAAGAGCAAA GAACACTGGCCAAACCGGTGGATGCATTCCAATTTCCATTCAACCCTGATTCAGGTATCCCGAGAAAGGCCCCAC CGACAGAATCTCGAAAGCGGTCAGGGGGGGTGGAGCAGGATGTATTCTCTGATTATGAAGAGGTCATCCAACCAC CGACTCCTCTCCAGCATCATCCACGCACCCATCGTCGCACACTGTTCAAGGAGAGGTGTACGCCATTTTACGGAG TTCCGAATCGCCGGGTTGCATTCGAGGTCTGCAAAGGCCTCGTTCGTGGGCTGGATCTTTTGCGACAAGCAGGAT TTTTACATCGGAATATCAGCGGCTCCAGCTGCGTGTTCTACTACGACAAGGATGCCCAAGCCTATCAGGCTAAAC TGATCGAGTTGGAGTATTGCAAAAGATATCAAGATACACGTTTCCATGATCCCAAATCGGTTCCACGGGAATTTG CCGCTGTTGAAGTCATGGATTCCCAGTGGAACCTCGCGAAGACCGGTCTGGATGTCCTCCAGTTGGATGAATTAC TCCCCCCTTTCCACCGACATTATTACCACGATCTGGAGTCGCTCTTCTGGCTATTAACCTGGTACACAACCACGC ATTTGCCTATCGACAGGGATGAAAATCAGGAAATCGTGGCTAATCTTGACCTTGTCAGTTGGAAGACAAAGGTCT TTGATGTTTTGTTCCCGCGGGAATCCCATTTAGAGCGCCGCCGAAACCTATGGGAAAGACCAAACCAGGTTTACC GTGACTTGAAGGCTGCCGTCGGATGGCCAATAGACACGGTCACTATCCTCCTCAGCCTCAGCAAGATACCTTCTT ACTTTGATTCGGAATACACGGCACTTTATCGGAATCCTCCACAGGATAAAGCAGTGCGGTGGCCCGATGCAAGGT TCAACGATTCACTATACAAGAAATTTATCAACACTCTGGACGATGTCGCTGCTCGTCTTGGAACAGTTGGTTCTG TTTCAATGTGGGGTTTACTGAATGAGGGGAGGATGAAGAACAAGAGGCCCGGGAAAGAAAAGGACGAGACTGACA GAATCCATAAGAAGCGGGGATGAACAGTGGAGGTTAGCGACAGTCGGAGTCGGAAACGGTGAAGTAATAGGCGCC CGTTCGTGTCCCAAAGTCCGCCTGATTAATTATGCATCGTGGCGGCGATATATTGTTACTGGCTGTCTTTACTTG GAAATGTGACTAATGCCCACTGTAGTCCCCAAAGTCCGTCTGGGCTGACGGCAAGACGCGCATTATTACTGTATT AGCAGAATATCATACTGCCATTTGAAATTGG |
| Length | 2656 |
Gene
| Sequence id | CopciAB_420995.T0 |
|---|---|
| Sequence |
>CopciAB_420995.T0 ACCCACACAGACCCACGATTCTCCTGCACGTACCCTCGCCTTCCACTCGTTATCCTCGCTCTCTCTCTTGAACTA TATCTACAAGAATGGAGACAGAACAGTTAAAAGCATCAGAACAGGTGAGCTATTATCCCATCCCCCTCCCATCCA TCGATTCTCCCAATGGCCTCGGAATTGAAACCGAATTTAACTACTGAATCACCTCCAAAGGACCAAAACTACTTC GACAGTCGCACTGCTGCGCAAAAGGACGCAGAGGAACAGCGCAAAACCCATCTTTTCAATGTGAAAGATCACCTT TTACAGTGTGACTTTTCCACCTGGTTCGAAACCTGCCTTTCGAATCCTGACGATATGAAGTGGAGCGAGAAATCT GCTTTGGATGTCAAGAGGAGACTTCTTACATTGTCGCCGCCCGTCCTCACAGACGAAGGCTGGGTGCCATTGAAG AAGGCCTTGGGTGAAACCGACAAGAAGGAGGACACCGTTTACGCATCTATGACGGGTATCATTTCGAAAATCAAG CATGCGGCGGCTCAGGAACTTAAGCTTGATTCCGATTTTGAGGGACAAATAACAGTCTATTCCTCCACTCCCACT CGACCAACTACTAACGACGTCCATGGACCCAACTTGAAGCCTGACGGTCGCTTTGTACTCGCACCACCGCCGCCT CAGCCTCCAACTCATGTTCTCTACCGCCGATCATCGCGGCTAGCGAAAAAGATGGAGGAGGAGGCGCGTAGTCAG AGGAATGCCACTGCAAGCAGCAACGTTTCCGACATATCCATGTCTACGTCTGATTCGGTTGCGCTCTTTGAATAC AAGCTCAAGGAGGCAGATCAGCGACAGGTGGGTCGTTGGTTCCTCAGATTACCAGGCCCAAAGGCTTATTGAGTC ATCGCAACAGAACTACCGGCAAATGCTGGGAGGTGCAGCTTTCGTGCTGTACAACGACCCACGTCGCACTCGGGT CTATGGGGTCACCATTGAAGAAACTACCACGCGGCTGTGGCACTTTGATCGCGGAGCGGTCCTCTTCAGCCCTAC GTTTGACTGTCAAGAGGTAGACTTTACGGTTTATTCATCTTTTCCAAGCTGACATCTATCCAAGAAACCTGAATA CCTCATCAGGATCTGCTTGTATCTGTCATACGCGAGCCCGATACAGCTTGGCTACGACCCCACCGTGAAGAAGGT CGAGAGCAAAAATGGGAAAGGGCATTCACCAGACTTCATCTACCAAGTCAACAACAAGTACTATCGAACCATCGG ACAGCCACTGTCTGAAACAAGTGCATTCTGGACAATCTCGAGAGGAATAAGGACTTGGGTCGTCCAGGAGTGCGA TGAGAATGGCGCGATCAAGCCAAATATAGCCGAGGCCGTTCTAAGGGACCTATGGCTGTATGAAGATGAGCCTGA CGAAAAGGCTATTCAGACTGAGATTCTCAGTCACTTATCCAGACGCAACATTCCAGTCAAGGCGATGGAACAGCT GTTCGTCCATATATTGGAGGATGTCACCCTTTCAGGGAAAGAGCAAAGAACACTGGCCAAACCGGTGGATGCATT CCAATTTCCATTCAACCCTGATTCAGGTATCCCGAGAAAGGCCCCACCGACAGAATCTCGAAAGCGGTCAGGGGG GGTGGAGCAGGATGTATTCTCTGATTATGAAGAGGTCATCCAACCACCGACTCCTCTCCAGCATCATCCACGCAC CCATCGTCGCACACTGTTCAAGGAGAGGTGTACGCCATTTTACGGAGTTCCGAATCGCCGGGTTGCATTCGAGGT CTGCAAAGGCCTCGTTCGTGGTGCGCGATGAGCTCTCACTCACCCTTGCTTACAAGACTGACAGCACAGCCAGGG CTGGATCTTTTGCGACAAGCAGGATTTTTACATCGGAATATCAGCGGCTCCAGCTGCGTGTTCTACTACGACAAG GATGCCCAAGCCTATCAGGCTAAACTGATCGAGTTGGAGTATTGCAAAAGATATCAAGATACACGTTTCCATGAT CCCAAATCGGTATGTTTCGTATGTTCTTCCACGACTTGCAGCTTTCTGACCATTGGTCAAATGCAGGTTCCACGG GAATTTGCCGCTGTTGAAGTCATGGATTCCCAGTGGAACCTCGCGAAGACCGGTCTGGATGTCCTCCAGTTGGAT GAATTACTCCCCCCTTTCCACCGACATTATTACCACGATCTGGAGTCGCTCTTCTGGCTATTAACCTGGTACACA ACCACGCATTTGCCTATCGACAGGGATGAAAATCAGGAAATCGTGGCTAATCTTGACCTTGTCAGTTGGAAGACA AAGGTCTTTGATGTTTTGTTCCCGCGGGAATCCCATTTAGAGCGCCGCCGAAACCTATGGGAAAGACCAAACCAG GTTTACCGTGACTTGAAGGCTGCCGTCGGATGGCCAATAGACACGGTCACTATCCTCCTCAGCCTCAGCAAGATA CCTTCTTACTTTGATTCGGAATACACGGCACTTTATCGGAATCCTCCACAGGATAAAGCAGTGCGGTGGCCCGAT GCAAGGTTCAACGATTCACTATACAAGAAATTTATCAACACTCTGGACGATGTCGCTGCTCGTCTTGGAACAGTT GGTTCTGTTTCAATGTGGGGTTTACTGAATGAGGGGAGGATGAAGAACAAGAGGCCCGGGAAAGAAAAGGACGAG ACTGACAGAATCCATAAGAAGCGGGGATGAACAGTGGAGGTTAGCGACAGTCGGAGTCGGAAACGGTGAAGTAAT AGGCGCCCGTTCGTGTCCCAAAGTCCGCCTGATTAATTATGCATCGTGGCGGCGATATATTGTTACTGGCTGTCT TTACTTGGAAATGTGACTAATGCCCACTGTAGTCCCCAAAGTCCGTCTGGGCTGACGGCAAGACGCGCATTATTA CTGTATTAGCAGAATATCATACTGCCATTTGAAATTGG |
| Length | 2963 |