CopciAB_421651
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_421651 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | NRC2 | Synonyms | 421651 |
| Uniprot id | Functional description | Serine/Threonine protein kinases, catalytic domain | |
| Location | scaffold_5:1324385..1327371 | Strand | - |
| Gene length (nt) | 2987 | Transcript length (nt) | 2285 |
| CDS length (nt) | 2091 | Protein length (aa) | 696 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN0144_nrc2 |
| Schizosaccharomyces pombe | ppk14 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7259525 | 69.1 | 4.562E-291 | 910 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB22507 | 67.1 | 9.058E-279 | 863 |
| Lentinula edodes NBRC 111202 | Lenedo1_1034753 | 65.4 | 1.278E-272 | 845 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1443391 | 65.3 | 1.401E-271 | 842 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_23126 | 63 | 1.775E-268 | 833 |
| Grifola frondosa | Grifr_OBZ77065 | 62.3 | 3.782E-258 | 803 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_68563 | 74.5 | 3.223E-238 | 745 |
| Flammulina velutipes | Flave_chr02AA00122 | 60.9 | 3.199E-231 | 725 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_41847 | 69.9 | 3.181E-231 | 725 |
| Schizophyllum commune H4-8 | Schco3_2623551 | 59.3 | 3.933E-231 | 725 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1039309 | 81.1 | 2.737E-212 | 670 |
| Pleurotus ostreatus PC9 | PleosPC9_1_49694 | 81.1 | 2.679E-212 | 670 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_99816 | 81.2 | 1.302E-210 | 665 |
| Auricularia subglabra | Aurde3_1_1273084 | 50.5 | 7.988E-196 | 623 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | NRC2 |
|---|---|
| Protein id | CopciAB_421651.T0 |
| Description | Serine/Threonine protein kinases, catalytic domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd05574 | STKc_phototropin_like | - | 331 | 641 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 335 | 617 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08286 |
EggNOG
| COG category | Description |
|---|---|
| T | Serine/Threonine protein kinases, catalytic domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
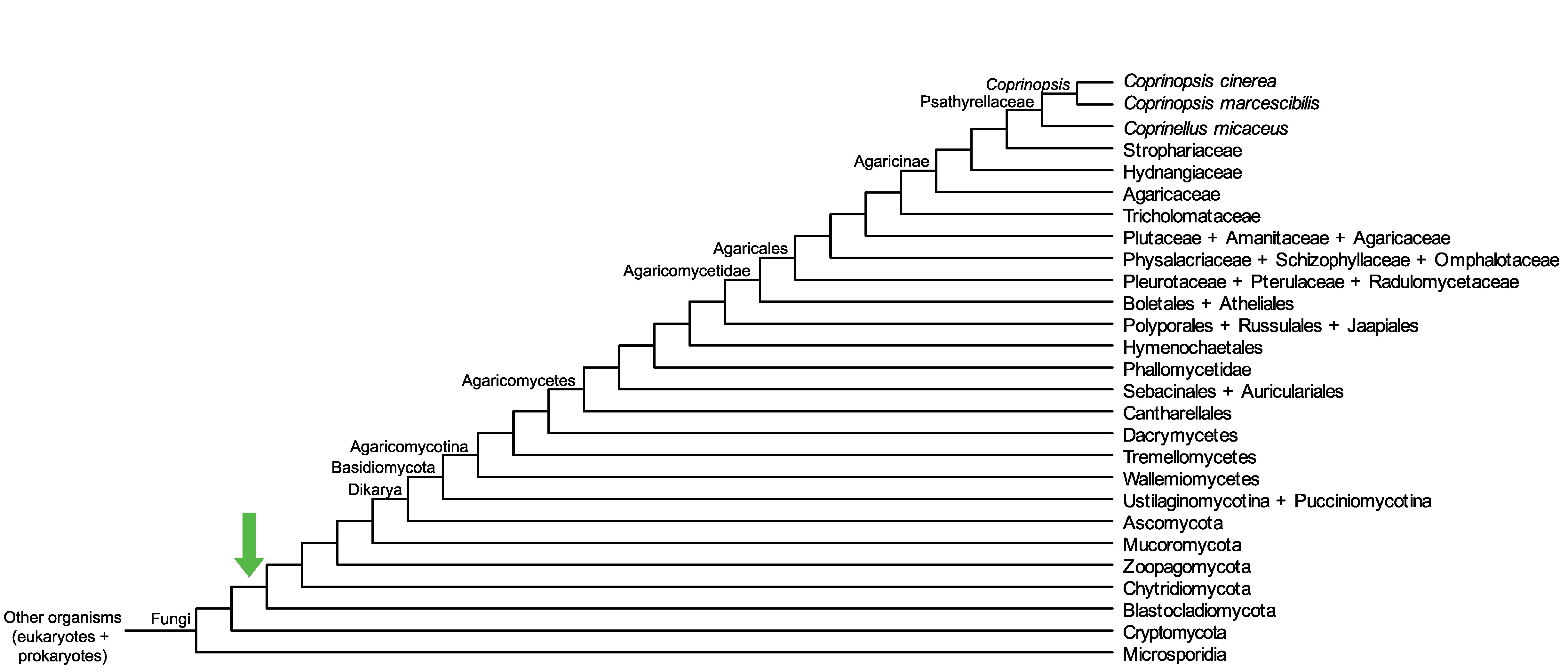
Conservation of CopciAB_421651 across fungi.
Arrow shows the origin of gene family containing CopciAB_421651.
Protein
| Sequence id | CopciAB_421651.T0 |
|---|---|
| Sequence |
>CopciAB_421651.T0 MPTNHQAAQYENLTVPSPKRPTEGHDSHSLPRSPSLSSWKSFLRFPNQSSKKPSNVPPSLNLDNRSVSGLSYNSA TSTPALATPALTPTSYLSDQRSSYNSSNTQSSDSNRPPAYQEPPSTTPRNVSYPLDHHHNAPSTDALAAATTAGT SQLKAPTYPGKQRSLPHPFEPGSSKSKPYTTTPSQSSFGAPPTTSRPRNNGPLTPKGVGASASRFLRRVASAPNA KGLFTLGSKSASTTKNGLLAPSESAPPLPTVSSSMEDGDSLDTASSAESSRATPRYGLRLTPPKTAPPMRDRLKS APSQPDPSKIPFRRTYSSNSIKVGRVEVGPSSFLKVKMLGKGDVGRVYLVREKKTNKLYAMKVLSKKEMIERKKI KRALTEQEILATANHPFIVTLYHSFQSEGYLYFCMEYCMGGEFFRALQTRPGKCLPEDGSRFYAAEVVAALEYLH LMGFIYRDLKPENILLHQSGHIMLSDFDLAKQSAEYGGLPSMVHSEQNGVPLVDTMSCTANFRTNSFVGTEEYIA PEVIAAQGHTAAVDWWTLGILIYEMIYATTPFKGQERNETFNNIRHQPVTFRDSPKISSAGKDCMTRLLDKNERT RLGSRSGASEVKQHKWFSKINWGLLRNTRPPIVPSSSNGLDAAHFRHMRESTSLHLDLEDQSKAQPSADLLADDG QPADVLFKAFNSITLHHDGDT |
| Length | 696 |
Coding
| Sequence id | CopciAB_421651.T0 |
|---|---|
| Sequence |
>CopciAB_421651.T0 ATGCCTACCAACCACCAGGCCGCACAGTATGAAAATCTGACAGTTCCCTCGCCCAAGAGGCCCACAGAGGGCCAC GATTCTCACTCCTTACCCCGCTCGCCCTCTCTTTCTTCCTGGAAATCCTTCCTTCGTTTCCCCAACCAATCTTCA AAGAAACCTTCCAATGTTCCACCAAGCCTAAACCTAGACAATCGTTCAGTATCAGGATTATCGTATAATAGTGCC ACAAGCACTCCGGCACTCGCAACTCCAGCACTAACCCCAACATCATACCTTTCGGACCAACGTTCTTCGTATAAT TCTTCCAATACCCAATCTAGTGACTCGAATAGACCTCCAGCCTATCAAGAACCACCCTCGACGACCCCGAGGAAT GTATCATATCCCCTGGACCACCACCACAATGCCCCGTCCACAGACGCGCTCGCAGCCGCCACGACAGCAGGCACT TCCCAGCTAAAGGCGCCCACATACCCAGGAAAACAACGTTCGCTTCCTCACCCCTTCGAACCCGGTTCATCCAAG TCGAAACCCTACACGACAACTCCTTCCCAGTCTTCCTTTGGTGCTCCTCCCACGACCTCGCGTCCACGAAACAAC GGGCCCCTGACCCCAAAGGGTGTAGGAGCTTCGGCTTCTCGCTTCCTTCGCAGAGTGGCTTCAGCACCCAACGCC AAGGGGCTGTTCACCCTAGGATCCAAGTCTGCATCCACCACGAAGAATGGTCTTCTAGCACCTTCCGAATCTGCA CCACCGCTACCGACAGTGTCCAGTTCGATGGAGGATGGTGATTCGCTAGACACCGCCTCTTCGGCAGAATCTTCA AGGGCTACCCCACGTTATGGGCTTCGATTGACACCGCCCAAGACAGCCCCGCCGATGAGAGATCGTTTGAAGAGT GCTCCATCTCAGCCCGACCCTTCAAAGATTCCGTTCAGACGGACGTATTCCAGCAACTCCATCAAAGTGGGACGG GTGGAGGTGGGCCCATCGAGCTTTTTGAAAGTGAAGATGCTGGGGAAGGGGGACGTTGGAAGGGTTTACCTGGTG CGGGAGAAGAAAACGAACAAACTCTACGCGATGAAAGTTTTGTCAAAGAAAGAGATGATTGAGAGGAAAAAGATT AAACGGGCTCTCACGGAGCAAGAAATCCTTGCAACCGCGAACCACCCTTTCATCGTTACCCTATATCACTCATTT CAGTCTGAAGGCTATTTATATTTCTGCATGGAATATTGCATGGGAGGGGAGTTCTTCCGCGCGCTGCAGACACGC CCGGGCAAGTGCTTGCCCGAAGATGGCTCGCGATTTTATGCCGCGGAGGTGGTTGCTGCGTTGGAATATCTTCAT CTCATGGGCTTCATTTATCGCGATTTGAAGCCCGAAAATATTCTTTTGCACCAATCAGGGCACATCATGCTGTCC GACTTTGACCTGGCGAAGCAGTCTGCAGAGTACGGGGGCCTACCTTCGATGGTTCACTCTGAGCAGAACGGGGTT CCTCTGGTGGATACGATGTCATGTACAGCGAACTTTAGGACGAACTCTTTCGTTGGCACGGAGGAGTACATTGCA CCAGAGGTGATCGCTGCGCAGGGCCATACAGCCGCTGTGGATTGGTGGACTTTAGGAATTCTCATTTATGAGATG ATTTATGCCACGACTCCGTTCAAAGGCCAGGAACGTAATGAGACCTTCAATAATATCCGCCATCAGCCCGTGACA TTCCGCGACTCACCAAAAATCTCATCAGCCGGCAAAGACTGCATGACTCGATTACTGGACAAGAACGAACGGACA AGACTAGGCAGTCGGAGCGGGGCGAGTGAGGTGAAGCAACACAAATGGTTCTCCAAGATCAATTGGGGCCTGTTG CGGAATACGCGGCCACCGATCGTGCCGTCGTCATCCAACGGTCTCGATGCAGCCCATTTCCGACATATGAGAGAA TCGACGTCGCTGCACCTCGATCTTGAGGATCAGAGCAAGGCGCAGCCTTCGGCAGATCTGCTTGCCGACGACGGG CAGCCTGCGGACGTGTTGTTCAAAGCGTTCAACAGTATAACGCTGCACCACGACGGGGACACATGA |
| Length | 2091 |
Transcript
| Sequence id | CopciAB_421651.T0 |
|---|---|
| Sequence |
>CopciAB_421651.T0 GTCCTCAATTCCTGTGTTTTCTCCCTCATTTTCGACATAATCGACGCCCAGATCATACTGGCTAATGCCTACCAA CCACCAGGCCGCACAGTATGAAAATCTGACAGTTCCCTCGCCCAAGAGGCCCACAGAGGGCCACGATTCTCACTC CTTACCCCGCTCGCCCTCTCTTTCTTCCTGGAAATCCTTCCTTCGTTTCCCCAACCAATCTTCAAAGAAACCTTC CAATGTTCCACCAAGCCTAAACCTAGACAATCGTTCAGTATCAGGATTATCGTATAATAGTGCCACAAGCACTCC GGCACTCGCAACTCCAGCACTAACCCCAACATCATACCTTTCGGACCAACGTTCTTCGTATAATTCTTCCAATAC CCAATCTAGTGACTCGAATAGACCTCCAGCCTATCAAGAACCACCCTCGACGACCCCGAGGAATGTATCATATCC CCTGGACCACCACCACAATGCCCCGTCCACAGACGCGCTCGCAGCCGCCACGACAGCAGGCACTTCCCAGCTAAA GGCGCCCACATACCCAGGAAAACAACGTTCGCTTCCTCACCCCTTCGAACCCGGTTCATCCAAGTCGAAACCCTA CACGACAACTCCTTCCCAGTCTTCCTTTGGTGCTCCTCCCACGACCTCGCGTCCACGAAACAACGGGCCCCTGAC CCCAAAGGGTGTAGGAGCTTCGGCTTCTCGCTTCCTTCGCAGAGTGGCTTCAGCACCCAACGCCAAGGGGCTGTT CACCCTAGGATCCAAGTCTGCATCCACCACGAAGAATGGTCTTCTAGCACCTTCCGAATCTGCACCACCGCTACC GACAGTGTCCAGTTCGATGGAGGATGGTGATTCGCTAGACACCGCCTCTTCGGCAGAATCTTCAAGGGCTACCCC ACGTTATGGGCTTCGATTGACACCGCCCAAGACAGCCCCGCCGATGAGAGATCGTTTGAAGAGTGCTCCATCTCA GCCCGACCCTTCAAAGATTCCGTTCAGACGGACGTATTCCAGCAACTCCATCAAAGTGGGACGGGTGGAGGTGGG CCCATCGAGCTTTTTGAAAGTGAAGATGCTGGGGAAGGGGGACGTTGGAAGGGTTTACCTGGTGCGGGAGAAGAA AACGAACAAACTCTACGCGATGAAAGTTTTGTCAAAGAAAGAGATGATTGAGAGGAAAAAGATTAAACGGGCTCT CACGGAGCAAGAAATCCTTGCAACCGCGAACCACCCTTTCATCGTTACCCTATATCACTCATTTCAGTCTGAAGG CTATTTATATTTCTGCATGGAATATTGCATGGGAGGGGAGTTCTTCCGCGCGCTGCAGACACGCCCGGGCAAGTG CTTGCCCGAAGATGGCTCGCGATTTTATGCCGCGGAGGTGGTTGCTGCGTTGGAATATCTTCATCTCATGGGCTT CATTTATCGCGATTTGAAGCCCGAAAATATTCTTTTGCACCAATCAGGGCACATCATGCTGTCCGACTTTGACCT GGCGAAGCAGTCTGCAGAGTACGGGGGCCTACCTTCGATGGTTCACTCTGAGCAGAACGGGGTTCCTCTGGTGGA TACGATGTCATGTACAGCGAACTTTAGGACGAACTCTTTCGTTGGCACGGAGGAGTACATTGCACCAGAGGTGAT CGCTGCGCAGGGCCATACAGCCGCTGTGGATTGGTGGACTTTAGGAATTCTCATTTATGAGATGATTTATGCCAC GACTCCGTTCAAAGGCCAGGAACGTAATGAGACCTTCAATAATATCCGCCATCAGCCCGTGACATTCCGCGACTC ACCAAAAATCTCATCAGCCGGCAAAGACTGCATGACTCGATTACTGGACAAGAACGAACGGACAAGACTAGGCAG TCGGAGCGGGGCGAGTGAGGTGAAGCAACACAAATGGTTCTCCAAGATCAATTGGGGCCTGTTGCGGAATACGCG GCCACCGATCGTGCCGTCGTCATCCAACGGTCTCGATGCAGCCCATTTCCGACATATGAGAGAATCGACGTCGCT GCACCTCGATCTTGAGGATCAGAGCAAGGCGCAGCCTTCGGCAGATCTGCTTGCCGACGACGGGCAGCCTGCGGA CGTGTTGTTCAAAGCGTTCAACAGTATAACGCTGCACCACGACGGGGACACATGAGTTGTGAGAGACGTGCACAC AGGGTGTGTGGCATACCTCGGAGGGGAGTCGGGATTTCGGTGATTGCGGTAGGATTGTGGAATCTTTTATCTCGT GTGGTACTTAGGCAGTAATAATTTTGCGAAAGCGG |
| Length | 2285 |
Gene
| Sequence id | CopciAB_421651.T0 |
|---|---|
| Sequence |
>CopciAB_421651.T0 GTCCTCAATTCCTGTGTTTTCTCCCTCATTTTCGACATAATCGACGCCCAGATCATACTGGCTAATGCCTACCAA CCACCAGGCCGCACAGTATGAAAATCTGACAGTTCCCTCGCCCAAGAGGCCCACAGAGGGCCACGATTCTCACTC CTTACCCCGCTCGCCCTCTCTTTCTTCCTGGAAATCCTTCCTTCGTTTCCCCAACCAATCTTCAAAGAAACCTTC CAATGTTCCACCAAGCCTAAACCTAGACAATCGTTCAGTATCAGGATTATCGTATAATAGTGCCACAAGCACTCC GGCACTCGCAACTCCAGCACTAACCCCAACATCATACCTTTCGGACCAACGTTCTTCGTATAATTCTTCCAATAC CCAATCTAGTGACTCGAATAGACCTCCAGCCTATCAAGAACCACCCTCGACGACCCCGAGGAATGTATCATATCC CCTGGACCACCACCACAATGCCCCGTCCACAGACGCGCTCGCAGCCGCCACGACAGCAGGCACTTCCCAGCTAAA GGCGCCCACATACCCAGGAAAACAACGTTCGCTTCCTCACCCCTTCGAACCCGGTTCATCCAAGTCGAAACCCTA CACGACAACTCCTTCCCAGTCTTCCTTTGGTGCTCCTCCCACGACCTCGCGTCCACGAAACAACGGGCCCCTGAC CCCAAAGGGTGTAGGAGCTTCGGCTTCTCGCTTCCTTCGCAGAGTGGCTTCAGCACCCAACGCCAAGGGGCTGTT CACCCTAGGATCCAAGTCTGCATCCACCACGAAGAATGGTCTTCTAGCACCTTCCGAATCTGCACCACCGCTACC GACAGTGTCCAGTTCGATGGAGGATGGTGATTCGCTAGACACCGCCTCTTCGGCAGAATCTTCAAGGGCTACCCC ACGTTATGGGCTTCGATTGACACCGCCCAAGACAGCCCCGCCGATGAGAGATCGTTTGAAGAGTGCTCCATCTCA GCCCGACCCTTCAAAGATTCCGTTCAGACGGACGTATTCCAGCAACTCCATCAAAGTGGGACGGGTGAGTTCGTA CCTGCTTTGCATTCTTGATACCGCACTCATCATCCGTGTTTTCAGGTGGAGGTGGGCCCATCGAGCTTTTTGAAA GTGAAGATGCTGGGGAAGGGGGACGTTGGAAGGGTTTACCTGGTGCGGGAGAAGAAAACGAACAAACTCTACGCG ATGAAAGGTAAAGGTGTTCAAACTCATCGGTGGCCTCTTGGGCCCCGGTCGTCTGTTTAGCGACGTTAAGCTGAC TTGGCCATCTGATCCGCATTCACAGTTTTGTCAAAGAAAGAGATGATTGAGAGGAAAAAGATTAAACGGGCTCTC ACGGAGCAAGAAATCCTTGCAACCGCGAACCACCCTTTCATCGTTACCCTATATCACTCATTTCAGTCTGAAGGC TATTTATATTTCTGCATGGAATATTGCATGGGAGGGGAGTTCTTCCGCGCGCTGCAGACACGCCCGGGCAAGTGC TTGCCCGAAGATGGCTCGCGATTTTATGCCGCGGAGGTGGTTGCTGCGTTGGAATATCTTCATCTCATGGGCTTC ATTTATCGCGATTTGAAGCCCGAAAGTATGTAGCCTTTCCTCTTCATGGTTTCACCGGTGCTGACCCAGACCCTA CTGTAGATATTCTTTTGCACCAATCAGGGCACATCATGCTGTCCGACTTTGACCTGGCGAAGCAGTCTGCAGAGT ACGGGGGCCTACCTTCGATGGTTCACTCTGAGCAGAACGGGGTATGTAGCTGGAGCGTCGAGATGGAACCCTGCC CGTTTGGCTCACTTGCTCTTTGTTTAGGTTCCTCTGGTGGATACGATGTCATGTACAGCGAACTTTAGGACGAAC TCTTTCGTTGGCACGGAGGGTAAGAGGTTTTCTCTTTGATTACCGCCTGACCAGGTGGTTTACACAGCGATTCCA GAGTACATTGCACCAGAGGTGATCGCTGCGCAGGGCCATACAGCCGCTGTGGATTGGTGGACTTTAGGAATTCTC ATTTATGAGATGATTGTACGTATATCTGGTGGAGTCTATCAAGATGGATCAGAGTTACTGACGTGTTTGGTGCCC TTTTGGGTGAACGACTACAGTATGCCACGACTCCGTTCAAAGGCCAGGAACGTAATGAGACCTTCAATAATATCC GCCATCAGCCCGTGACATTCCGCGACTCACCAAAAATCTCATCGTAAGTTCTGGGCTGTTTGCTTTGCTCTCGTT TCTTTCATCGTTTCGACGACTGATGCGCGGACTTTGCGTTCATGGGCGTCGCGCATCATCGCGATCCGATTTGGG AGATTTCTGTTTAGGAATAACCCCTGCCTAATCCGATCAACTGAGCTGACTATTACTCACAGAGCCGGCAAAGAC TGCATGACTCGATTACTGGACAAGAACGAACGGACAAGACTAGGCAGTCGGAGCGGGGCGAGTGAGGTGAAGCAA CACAAATGGTTCTCCAAGATCAATTGGGGCCTGTTGCGGAATACGCGGCCACCGGTGAGTAGTCTCCCTCTGAGC ATTCGACGGAGAGAAATGGCGGGGGCATGAGGGAAGCCTTCGCGCATTCATTCATTCCTATTGGGAGTTTCTTGC CTTCATTGGACCTGTACTGATAAATTTGCACCAGATCGTGCCGTCGTCATCCAACGGTCTCGATGCAGCCCATTT CCGACATATGAGAGAATCGACGTCGCTGCACCTCGATCTTGAGGATCAGAGCAAGGCGCAGCCTTCGGCAGATCT GCTTGCCGACGACGGGCAGCCTGCGGACGTGTTGTTCAAAGCGTTCAACAGTATAACGCTGCACCACGACGGGGA CACATGAGTTGTGAGAGACGTGCACACAGGGTGTGTGGCATACCTCGGAGGGGAGTCGGGATTTCGGTGATTGCG GTAGGATTGTGGAATCTTTTATCTCGTGTGGTACTTAGGCAGTAATAATTTTGCGAAAGCGG |
| Length | 2987 |