CopciAB_422799
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_422799 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | ENV7 | Synonyms | 422799 |
| Uniprot id | Functional description | Belongs to the protein kinase superfamily | |
| Location | scaffold_3:1624575..1626386 | Strand | + |
| Gene length (nt) | 1812 | Transcript length (nt) | 1346 |
| CDS length (nt) | 1242 | Protein length (aa) | 413 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | stk-9 |
| Aspergillus nidulans | AN10193_env7 |
| Saccharomyces cerevisiae | YPL236C_ENV7 |
| Schizosaccharomyces pombe | ppk13 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7266832 | 82 | 3.562E-228 | 701 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25751 | 75.8 | 4.142E-210 | 649 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191128 | 72.7 | 1.682E-206 | 638 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1454350 | 74.3 | 2.29E-206 | 638 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1094292 | 74.5 | 1.947E-206 | 638 |
| Schizophyllum commune H4-8 | Schco3_2627502 | 74.3 | 2.579E-206 | 638 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114902 | 72.4 | 3.891E-206 | 637 |
| Flammulina velutipes | Flave_chr09AA00959 | 73.3 | 3.595E-200 | 620 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_58850 | 70.4 | 4.275E-198 | 614 |
| Pleurotus ostreatus PC9 | PleosPC9_1_116091 | 71.9 | 2.516E-194 | 603 |
| Lentinula edodes B17 | Lened_B_1_1_5284 | 61.4 | 2.179E-177 | 554 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_12481 | 61.4 | 2.733E-177 | 554 |
| Lentinula edodes NBRC 111202 | Lenedo1_1072387 | 61 | 1.481E-175 | 549 |
| Auricularia subglabra | Aurde3_1_1331221 | 59.1 | 4.284E-150 | 476 |
| Grifola frondosa | Grifr_OBZ72390 | 59.2 | 7.053E-108 | 353 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | ENV7 |
|---|---|
| Protein id | CopciAB_422799.T0 |
| Description | Belongs to the protein kinase superfamily |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 43 | 168 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 257 | 402 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017441 | Protein kinase, ATP binding site |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08856 |
EggNOG
| COG category | Description |
|---|---|
| I | Belongs to the protein kinase superfamily |
| K | Belongs to the protein kinase superfamily |
| T | Belongs to the protein kinase superfamily |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
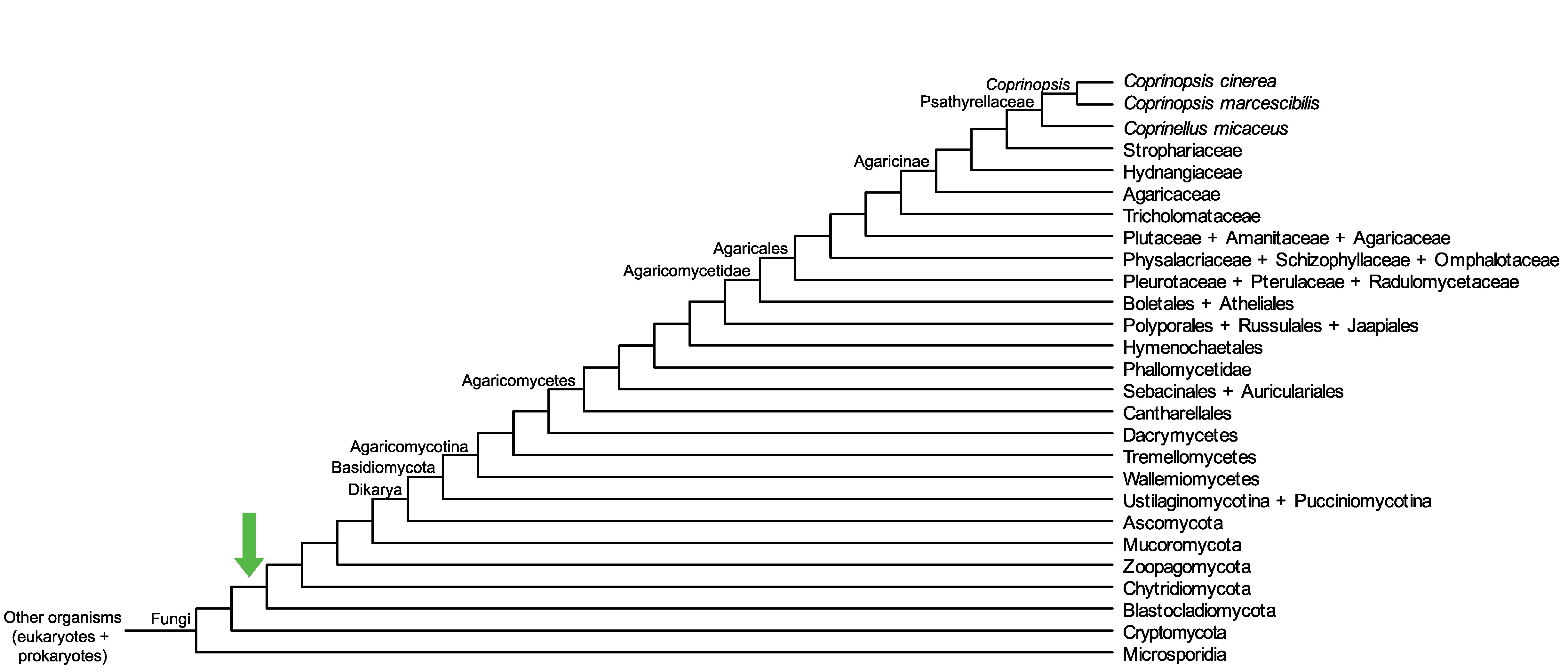
Conservation of CopciAB_422799 across fungi.
Arrow shows the origin of gene family containing CopciAB_422799.
Protein
| Sequence id | CopciAB_422799.T0 |
|---|---|
| Sequence |
>CopciAB_422799.T0 MALPPQLFRALETLKDQAKDALWAVSSCLCQQQPKLKINGRTFQIIKVLGEGGFSFVYLAQDEHSGRQFALKKIR CPTGSEGVKEAMREVEAYRRFKHPNIIRILDSAVVQDPEGEGQIVYLFLPLYKRGNLQDAINANLVNNRHFSEQD MLRLFRGTCLAVRAMHEYRPTSARSAIRTDPQRSNSTNSGPADDDDDTRFPRPEGDAEGGYSYDTPSVPLMGKQR ELVQNDVVFDGDEESELPQPQEGTPLDVVPYAHRDLKPGNIMIADDGKTPILMDFGSTMKAKIPIENRQQALLQQ DIAAEQSTMAYRAPELFDVKTGITLDEKVDIWSLGCTLYALAYSHSPFENTQTTEQGGSIAMAVLNAQYKHPQSA YSQGLRSLIDAMLKPNPQERPDINQVIELTDRVLQSLA |
| Length | 413 |
Coding
| Sequence id | CopciAB_422799.T0 |
|---|---|
| Sequence |
>CopciAB_422799.T0 ATGGCTCTACCTCCGCAACTATTTCGCGCTCTCGAAACGCTCAAAGATCAGGCCAAGGATGCACTTTGGGCCGTG TCGTCGTGTTTGTGCCAGCAGCAACCGAAACTCAAGATCAACGGACGGACATTCCAAATAATCAAGGTGCTAGGG GAAGGAGGCTTCTCCTTTGTGTATCTAGCTCAGGATGAACACAGTGGGCGACAATTCGCGCTCAAGAAGATTCGT TGCCCAACTGGCTCTGAAGGGGTGAAAGAAGCGATGCGAGAAGTTGAAGCTTATCGTAGATTCAAGCACCCCAAC ATCATCCGCATTCTGGACTCCGCAGTAGTCCAGGATCCCGAGGGCGAGGGACAGATCGTGTATCTCTTCCTACCG TTGTACAAGCGTGGCAACCTCCAAGACGCCATAAACGCCAACTTGGTGAACAATCGACACTTCTCCGAACAAGAC ATGCTCCGCCTGTTCCGTGGAACATGTCTCGCAGTACGGGCCATGCACGAATACCGACCAACGTCCGCTCGTTCC GCTATCCGCACCGATCCTCAGCGGTCGAATTCCACTAATTCTGGGCCTGCTGACGACGACGATGATACGCGCTTC CCTCGGCCAGAAGGTGACGCAGAGGGAGGCTACTCCTATGATACACCATCTGTTCCTCTCATGGGCAAACAACGA GAACTGGTGCAGAATGACGTCGTATTCGACGGCGACGAGGAGTCCGAATTGCCGCAACCACAAGAGGGTACCCCG CTGGACGTGGTCCCCTACGCACATCGGGATCTGAAACCTGGAAATATCATGATTGCAGATGACGGAAAAACACCT ATTCTCATGGACTTTGGAAGTACAATGAAAGCCAAAATACCCATAGAAAACCGGCAACAGGCCTTGCTGCAACAG GACATCGCCGCTGAGCAAAGCACGATGGCCTACCGCGCACCGGAACTTTTCGACGTCAAAACAGGCATTACCCTT GACGAAAAGGTCGACATCTGGTCTCTTGGGTGCACGCTATACGCATTGGCTTACTCTCATTCGCCGTTCGAAAAC ACCCAGACGACGGAACAGGGCGGCTCGATTGCGATGGCAGTGCTAAACGCGCAGTACAAGCACCCGCAATCGGCT TATTCGCAGGGGCTGCGGAGCCTCATCGATGCCATGTTGAAGCCAAATCCTCAAGAACGACCGGATATCAATCAG GTTATTGAATTGACGGATCGAGTTCTTCAATCGTTAGCGTAA |
| Length | 1242 |
Transcript
| Sequence id | CopciAB_422799.T0 |
|---|---|
| Sequence |
>CopciAB_422799.T0 ACCATTTCAACCACGAACACCTGTTATCAATGGCTCTACCTCCGCAACTATTTCGCGCTCTCGAAACGCTCAAAG ATCAGGCCAAGGATGCACTTTGGGCCGTGTCGTCGTGTTTGTGCCAGCAGCAACCGAAACTCAAGATCAACGGAC GGACATTCCAAATAATCAAGGTGCTAGGGGAAGGAGGCTTCTCCTTTGTGTATCTAGCTCAGGATGAACACAGTG GGCGACAATTCGCGCTCAAGAAGATTCGTTGCCCAACTGGCTCTGAAGGGGTGAAAGAAGCGATGCGAGAAGTTG AAGCTTATCGTAGATTCAAGCACCCCAACATCATCCGCATTCTGGACTCCGCAGTAGTCCAGGATCCCGAGGGCG AGGGACAGATCGTGTATCTCTTCCTACCGTTGTACAAGCGTGGCAACCTCCAAGACGCCATAAACGCCAACTTGG TGAACAATCGACACTTCTCCGAACAAGACATGCTCCGCCTGTTCCGTGGAACATGTCTCGCAGTACGGGCCATGC ACGAATACCGACCAACGTCCGCTCGTTCCGCTATCCGCACCGATCCTCAGCGGTCGAATTCCACTAATTCTGGGC CTGCTGACGACGACGATGATACGCGCTTCCCTCGGCCAGAAGGTGACGCAGAGGGAGGCTACTCCTATGATACAC CATCTGTTCCTCTCATGGGCAAACAACGAGAACTGGTGCAGAATGACGTCGTATTCGACGGCGACGAGGAGTCCG AATTGCCGCAACCACAAGAGGGTACCCCGCTGGACGTGGTCCCCTACGCACATCGGGATCTGAAACCTGGAAATA TCATGATTGCAGATGACGGAAAAACACCTATTCTCATGGACTTTGGAAGTACAATGAAAGCCAAAATACCCATAG AAAACCGGCAACAGGCCTTGCTGCAACAGGACATCGCCGCTGAGCAAAGCACGATGGCCTACCGCGCACCGGAAC TTTTCGACGTCAAAACAGGCATTACCCTTGACGAAAAGGTCGACATCTGGTCTCTTGGGTGCACGCTATACGCAT TGGCTTACTCTCATTCGCCGTTCGAAAACACCCAGACGACGGAACAGGGCGGCTCGATTGCGATGGCAGTGCTAA ACGCGCAGTACAAGCACCCGCAATCGGCTTATTCGCAGGGGCTGCGGAGCCTCATCGATGCCATGTTGAAGCCAA ATCCTCAAGAACGACCGGATATCAATCAGGTTATTGAATTGACGGATCGAGTTCTTCAATCGTTAGCGTAAATTA GACGTTGTTCTTCGTGAAAGGACGGATCTATTGGCATCTTTGCTAACTTATCATTATGTTTTATTACCTTC |
| Length | 1346 |
Gene
| Sequence id | CopciAB_422799.T0 |
|---|---|
| Sequence |
>CopciAB_422799.T0 ACCATTTCAACCACGAACACCTGTTATCAATGGCTCTACCTCCGCAACTATTTCGCGCTCTCGAAACGCTCAAAG ATCAGGCCAAGGATGCACTTTGGGCCGTGTCGTCGTGTTTGTGCCAGCAGCAACCGAAACTCAAGATCAACGGAC GGACATGTCAGTAACCAGTGTATTCGAGTGGTTGTCAGCTCATCCTCCGATTGTAGTCCAAATAATCAAGGTGCT AGGGGAAGGAGGCTTCTCCTTTGTGTATCTAGCTCAGGATGAACACAGTGGGGTACGTCGCTCCATGGTACAACC AGGCCGCAGGGTGTAATTGTGGTCTCTAGCGACAATTCGCGCTCAAGAAGATTCGTTGCCCAACTGGCTCTGAAG GGGTGAAAGAAGCGATGCGAGAAGTTGAAGCTTATCGTAGATTCAAGTGGGTCATTACTTTATTGAGCGACCGCC TCACTGAACGCCCTCCAGGCACCCCAACATCATCCGCATTCTGGTTAGTTGACCGATTCTCAATGGTACAAGGAC CATTTCTCACCAAAATGTTAGGACTCCGCAGTAGTCCAGGATCCCGAGGGCGAGGGACAGATCGTGTATCTCTTC CTACCGTTGTACAAGGTTCGTCCGATTCGTATGACCAGCTCAGGCATCACATATCCTCAGTCCTTGATAGCGTGG CAACCTCCAAGACGCCATAAACGCCAACTTGGTGAACAATCGACACTTCTCCGAACAAGACATGCTCCGCCTGTT CCGTGGAACATGTCTCGCAGTACGGGCCATGCACGAATACCGACCAACGTCCGCTCGTTCCGCTATCCGCACCGA TCCTCAGCGGTCGAATTCCACTAATTCTGGGCCTGCTGACGACGACGATGATACGCGCTTCCCTCGGCCAGAAGG TGACGCAGAGGGAGGCTACTCCTATGATACACCATCTGTTCCTCTCATGGGCAAACAACGAGAACTGGTGCAGAA TGACGTCGTATTCGACGGCGACGAGGAGTCCGAATTGCCGCAACCACAAGAGGGTACCCCGCTGGACGTGGTCCC CTACGCACATCGGGATCTGAAACCTGGGTTTGTGAACTCTGTCCTTTCAGGTCTCCATGGACTCATGTTTTTCAG AAATATCATGATTGCAGATGACGGAAAAACACCTATTCTCATGGACTTTGGAAGTACAATGAAAGCCAAAATACC CATAGAAAACCGGCAACAGGCCTTGCTGCAACAGGTTAGCCCAAGCCGCGGTTCTTAGCCTCGATGTGCCCTCTA ACCTTAAACCCCACCAAGGACATCGCCGCTGAGCAAAGCACGATGGCCTACCGCGCACCGGAACTTTTCGACGTC AAAACAGGCATTACCCTTGACGAAAAGGTCGACATCTGGGTGAGCCTTCTGTTCCATGGCAATGTACTTCTCTCA TATTCCTTGCAGTCTCTTGGGTGCACGCTATACGCATTGGCTTACTCTCATTCGCCGTTCGAAAACACCCAGACG ACGGAACAGGGCGGCTCGATTGCGATGGCAGTGCTAAACGCGCAGTACAAGCACCCGCAATCGGCTTATTCGCAG GGGCTGCGGAGCCTCATCGATGCCATGTTGAAGCCAAATCCTCAAGAACGACCGGATATCAATCAGGTAGGTCTA TCAAATTGATGTTTGCTTCTCTACAATTGAACTCGATGTCTCCAGGTTATTGAATTGACGGATCGAGTTCTTCAA TCGTTAGCGTAAATTAGACGTTGTTCTTCGTGAAAGGACGGATCTATTGGCATCTTTGCTAACTTATCATTATGT TTTATTACCTTC |
| Length | 1812 |