CopciAB_430560
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_430560 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | exgD | Synonyms | 430560 |
| Uniprot id | Functional description | Exo-beta-1,3-glucanase | |
| Location | scaffold_2:94460..97188 | Strand | + |
| Gene length (nt) | 2729 | Transcript length (nt) | 2246 |
| CDS length (nt) | 1983 | Protein length (aa) | 660 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7271152 | 60.1 | 2.272E-285 | 880 |
| Schizophyllum commune H4-8 | Schco3_2484696 | 64.4 | 3.296E-263 | 816 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_67883 | 53.5 | 1.723E-228 | 715 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_198709 | 53.3 | 3.971E-227 | 711 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | exgD |
|---|---|
| Protein id | CopciAB_430560.T0 |
| Description | Exo-beta-1,3-glucanase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 189 | 353 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 48 | 70 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01210 |
EggNOG
| COG category | Description |
|---|---|
| G | Exo-beta-1,3-glucanase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_9 |
Transcription factor
| Group |
|---|
| No records |
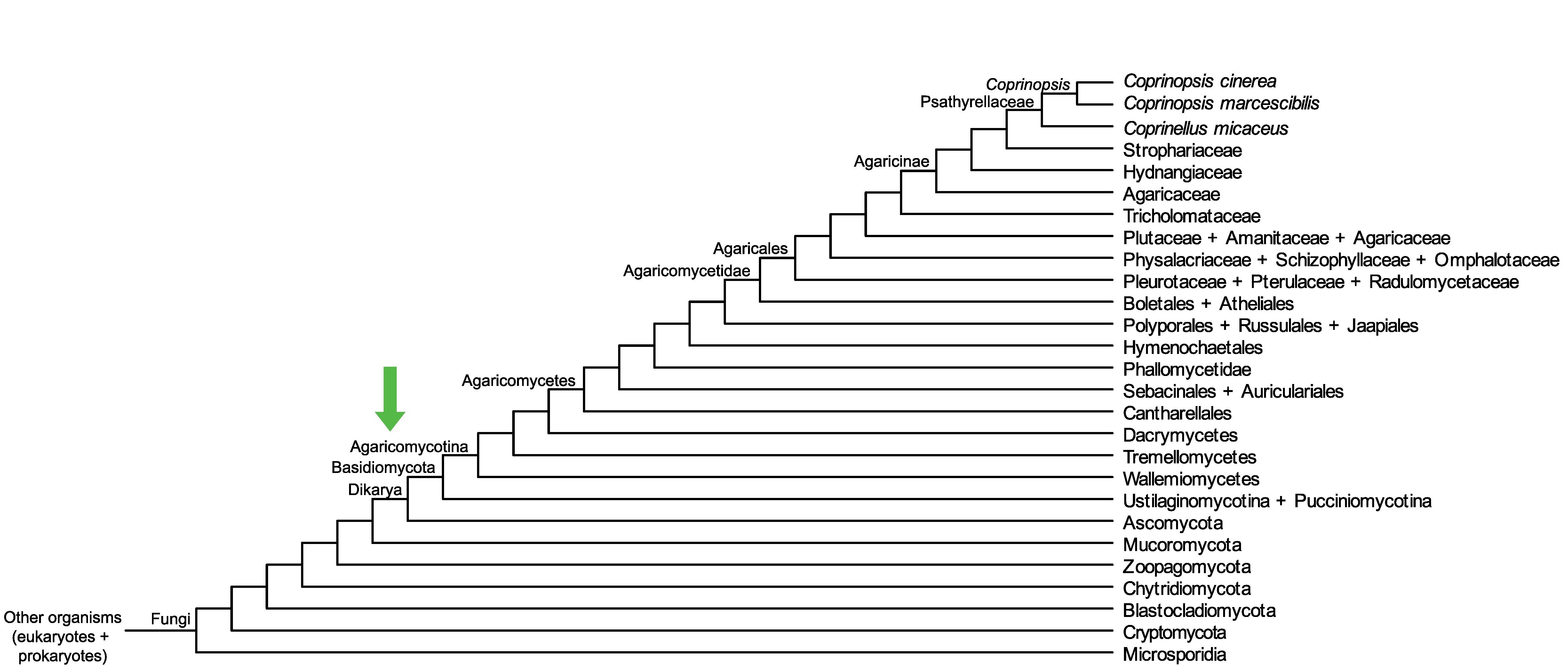
Conservation of CopciAB_430560 across fungi.
Arrow shows the origin of gene family containing CopciAB_430560.
Protein
| Sequence id | CopciAB_430560.T0 |
|---|---|
| Sequence |
>CopciAB_430560.T0 MEDTARAGSYSPFGTWEPSPRHSTGSKALLSEASSDGSVTKKPLHRRVWFIILFALTVIVVLFLAIFLPIYFTTI KHSDDSTSPSSRGTTGGNGSEISTEDGTKFIYLNPFGGHWVDDPDDPFNNDARPNSWTPPLNSSWKWGEDRIYGV NVGGLFVLEPFISPEIFQRYPGTKDEYELSEAMAADEANGGLSQLEEHYATFITEQDIAEIAGAGLNWIRVPIGF WAVETWEGEPFLERTSWKYFLRIIKWARKYGLRVALDLHAVPGSQNGYNHSGRLSQVNFLAGNMGIANAQRTLYT LRVFTEFISQPEYRDVIQVIELVNEPLAGEIGAEALSSFYLEAYNMIRKITGTGDGNGPYIAISDGLQPLSLWDG LLPGGDRVIMDGHPYFAFGGINTAPITEPSEDGKAGGTWPKAACDNWGPAFNHSRRTIGPTVGGEFSAAPNDCGL FLRGVGIESPHPQCPEYDDWENYNATMKEGLESFIMASFDALGDFFFWTWKIGPSIDGRVGTPLWSYKLGLENGW IPRDPRTSIGKCVAVGGNQEPFDGAYPPYRTGADPSPTIPASYSSQYPWPPASITEAAVPVELLPTYTNTAPIIT LPVELPTGTPTPLPSGLDGWFNDDDVEGGPAEINGCQYPDRYMAEFSTIPTAPCTGPTPA |
| Length | 660 |
Coding
| Sequence id | CopciAB_430560.T0 |
|---|---|
| Sequence |
>CopciAB_430560.T0 ATGGAAGACACAGCCCGTGCAGGCAGCTACTCTCCGTTTGGGACATGGGAACCTTCCCCTCGTCACTCGACTGGC AGCAAAGCGTTGCTATCTGAGGCATCAAGCGACGGATCCGTGACCAAGAAACCCCTTCATCGTCGTGTTTGGTTC ATCATCTTATTTGCTCTTACGGTCATTGTCGTTCTGTTTCTGGCCATCTTTCTTCCAATATACTTCACCACTATA AAGCACTCTGATGATAGTACTAGTCCATCTTCTCGAGGAACGACCGGAGGGAATGGTTCCGAGATATCGACGGAA GATGGGACGAAATTCATATATTTGAATCCCTTTGGAGGACACTGGGTGGATGATCCTGACGATCCGTTCAACAAC GACGCGAGGCCAAACTCCTGGACTCCGCCGCTAAACTCTTCGTGGAAGTGGGGAGAGGACCGAATCTACGGCGTC AACGTCGGTGGTCTCTTCGTTTTGGAGCCCTTCATCTCGCCAGAGATATTTCAGAGGTACCCAGGGACGAAGGAT GAGTATGAACTTTCAGAGGCCATGGCTGCAGACGAGGCCAACGGTGGATTATCCCAACTAGAAGAGCACTACGCC ACCTTCATTACAGAACAGGACATTGCCGAGATCGCAGGCGCTGGTCTAAATTGGATCAGGGTCCCCATTGGATTT TGGGCGGTTGAGACATGGGAAGGGGAACCATTTTTAGAGAGGACGTCGTGGAAATATTTCCTTCGGATAATCAAA TGGGCTAGAAAATACGGCCTGAGAGTGGCCCTGGATCTACATGCAGTCCCTGGATCGCAAAACGGCTACAATCAC TCGGGAAGATTATCTCAAGTTAACTTTCTGGCTGGGAACATGGGTATCGCCAATGCTCAGAGGACGCTCTATACC CTCCGCGTTTTTACTGAATTCATCTCCCAGCCTGAGTATAGAGACGTGATTCAAGTGATTGAGCTCGTAAATGAG CCTTTGGCTGGGGAGATAGGTGCAGAGGCCCTGTCGAGCTTCTACCTGGAAGCGTACAACATGATTAGGAAGATC ACAGGAACGGGAGACGGAAATGGGCCATACATCGCCATATCCGACGGTCTGCAACCACTCTCATTGTGGGATGGC CTCCTTCCTGGGGGTGACCGAGTTATCATGGACGGCCACCCATATTTCGCCTTCGGTGGCATAAACACAGCGCCC ATAACCGAACCCTCCGAAGATGGAAAGGCTGGAGGGACGTGGCCGAAAGCTGCGTGTGATAACTGGGGTCCCGCC TTCAACCACAGTCGAAGGACGATTGGCCCCACTGTTGGAGGAGAATTCTCGGCTGCACCCAATGATTGTGGCTTA TTCCTACGGGGTGTTGGTATCGAATCACCGCACCCTCAATGTCCGGAGTACGATGACTGGGAGAATTACAATGCC ACCATGAAAGAAGGATTGGAAAGCTTCATCATGGCCAGCTTCGACGCGTTGGGAGATTTCTTCTTCTGGACATGG AAGATCGGTCCTTCGATCGACGGTCGAGTAGGGACCCCCCTCTGGTCCTACAAGCTAGGACTCGAAAATGGTTGG ATACCCAGAGATCCTAGAACCTCGATCGGGAAATGTGTAGCTGTTGGGGGAAATCAAGAGCCTTTCGATGGAGCC TACCCACCATATCGGACGGGGGCTGATCCCTCACCCACCATTCCAGCCTCGTACTCCTCGCAATACCCTTGGCCA CCTGCATCCATCACAGAAGCCGCCGTTCCTGTAGAACTCCTCCCGACCTACACCAATACAGCACCGATCATTACC CTGCCAGTAGAGTTACCGACTGGGACCCCCACTCCTCTTCCCAGCGGGCTGGACGGCTGGTTCAACGACGACGAT GTGGAAGGAGGCCCAGCTGAGATTAATGGATGCCAGTACCCCGACCGCTATATGGCTGAATTCTCTACCATTCCC ACTGCACCTTGTACGGGTCCCACTCCCGCCTAA |
| Length | 1983 |
Transcript
| Sequence id | CopciAB_430560.T0 |
|---|---|
| Sequence |
>CopciAB_430560.T0 ACTGGTTCCTAAAAACCAACCCAAATCTGCCATTCAAGTCGTCGCTTCAGTAACTCAAGGGCTCGGGATCAGCGG CACATATGGAAGACACAGCCCGTGCAGGCAGCTACTCTCCGTTTGGGACATGGGAACCTTCCCCTCGTCACTCGA CTGGCAGCAAAGCGTTGCTATCTGAGGCATCAAGCGACGGATCCGTGACCAAGAAACCCCTTCATCGTCGTGTTT GGTTCATCATCTTATTTGCTCTTACGGTCATTGTCGTTCTGTTTCTGGCCATCTTTCTTCCAATATACTTCACCA CTATAAAGCACTCTGATGATAGTACTAGTCCATCTTCTCGAGGAACGACCGGAGGGAATGGTTCCGAGATATCGA CGGAAGATGGGACGAAATTCATATATTTGAATCCCTTTGGAGGACACTGGGTGGATGATCCTGACGATCCGTTCA ACAACGACGCGAGGCCAAACTCCTGGACTCCGCCGCTAAACTCTTCGTGGAAGTGGGGAGAGGACCGAATCTACG GCGTCAACGTCGGTGGTCTCTTCGTTTTGGAGCCCTTCATCTCGCCAGAGATATTTCAGAGGTACCCAGGGACGA AGGATGAGTATGAACTTTCAGAGGCCATGGCTGCAGACGAGGCCAACGGTGGATTATCCCAACTAGAAGAGCACT ACGCCACCTTCATTACAGAACAGGACATTGCCGAGATCGCAGGCGCTGGTCTAAATTGGATCAGGGTCCCCATTG GATTTTGGGCGGTTGAGACATGGGAAGGGGAACCATTTTTAGAGAGGACGTCGTGGAAATATTTCCTTCGGATAA TCAAATGGGCTAGAAAATACGGCCTGAGAGTGGCCCTGGATCTACATGCAGTCCCTGGATCGCAAAACGGCTACA ATCACTCGGGAAGATTATCTCAAGTTAACTTTCTGGCTGGGAACATGGGTATCGCCAATGCTCAGAGGACGCTCT ATACCCTCCGCGTTTTTACTGAATTCATCTCCCAGCCTGAGTATAGAGACGTGATTCAAGTGATTGAGCTCGTAA ATGAGCCTTTGGCTGGGGAGATAGGTGCAGAGGCCCTGTCGAGCTTCTACCTGGAAGCGTACAACATGATTAGGA AGATCACAGGAACGGGAGACGGAAATGGGCCATACATCGCCATATCCGACGGTCTGCAACCACTCTCATTGTGGG ATGGCCTCCTTCCTGGGGGTGACCGAGTTATCATGGACGGCCACCCATATTTCGCCTTCGGTGGCATAAACACAG CGCCCATAACCGAACCCTCCGAAGATGGAAAGGCTGGAGGGACGTGGCCGAAAGCTGCGTGTGATAACTGGGGTC CCGCCTTCAACCACAGTCGAAGGACGATTGGCCCCACTGTTGGAGGAGAATTCTCGGCTGCACCCAATGATTGTG GCTTATTCCTACGGGGTGTTGGTATCGAATCACCGCACCCTCAATGTCCGGAGTACGATGACTGGGAGAATTACA ATGCCACCATGAAAGAAGGATTGGAAAGCTTCATCATGGCCAGCTTCGACGCGTTGGGAGATTTCTTCTTCTGGA CATGGAAGATCGGTCCTTCGATCGACGGTCGAGTAGGGACCCCCCTCTGGTCCTACAAGCTAGGACTCGAAAATG GTTGGATACCCAGAGATCCTAGAACCTCGATCGGGAAATGTGTAGCTGTTGGGGGAAATCAAGAGCCTTTCGATG GAGCCTACCCACCATATCGGACGGGGGCTGATCCCTCACCCACCATTCCAGCCTCGTACTCCTCGCAATACCCTT GGCCACCTGCATCCATCACAGAAGCCGCCGTTCCTGTAGAACTCCTCCCGACCTACACCAATACAGCACCGATCA TTACCCTGCCAGTAGAGTTACCGACTGGGACCCCCACTCCTCTTCCCAGCGGGCTGGACGGCTGGTTCAACGACG ACGATGTGGAAGGAGGCCCAGCTGAGATTAATGGATGCCAGTACCCCGACCGCTATATGGCTGAATTCTCTACCA TTCCCACTGCACCTTGTACGGGTCCCACTCCCGCCTAAAACCCTCTCTCGCACACGATGTTGGGGTGAACTCTTA CAAGCCGAACTCTTACGGACGACAATTTTAATGAAACAGACCATATCTATTATATTGATAGTCTACTCCAGCAAT CTTACCTACTAGTTGTATGACAATTGTCTCGCTACAATGTGGATTCAGTTGCGCGAAGGCTCGCATTCCAC |
| Length | 2246 |
Gene
| Sequence id | CopciAB_430560.T0 |
|---|---|
| Sequence |
>CopciAB_430560.T0 ACTGGTTCCTAAAAACCAACCCAAATCTGCCATTCAAGTCGTCGCTTCAGTAACTCAAGGGCTCGGGATCAGCGG CACATATGGAAGACACAGCCCGTGCAGGCAGCTACTCTCCGTTTGGGACATGGGAACCTTCCCCTCGTCACTCGA CTGGCAGCAAAGCGTTGCTATCTGAGGCATCAAGCGACGGATCCGTGACCAAGAAACCCCTTCATCGTCGTGTTT GGTTCATCATCTTATTTGCTCTTACGGTCATTGTCGTTCTGTTTCTGGCCATCTTTCTTCCAATATACTTCACCA CTATAAAGCACTCTGATGATAGTACTAGTCCATCTTCTCGAGGAACGGTGCGTACCACTTTTTTGGAAGCACAGG GACTTTGGGACCTGACTACGATAGACCGGAGGGAATGGTTCCGAGATATCGACGGAAGATGGGACGAAATTCATA TATTTGAATCCCTTTGGAGGACACTGTGAGCGCTTCTTTTATCAAGCCTCGCTCTAACCGGCTGATCTTCGTTCC AAGGGGTGGATGATCCTGACGATCCGTTCAACAACGACGCGAGGCCAAACTCCTGGACTCCGCCGCTAAACTCTT CGTGGAAGTGGGGAGAGGACCGAATCTACGGCGTCAACGTCGGTGGTCTCTTCGTTTTGGAGCCCTTCATCTCGC CAGAGATATTTCAGAGGTACCCAGGGACGAAGGATGAGTATGAACTTTCAGAGGCCATGGCTGCAGACGAGGCCA ACGGTGGATTATCCCAACTAGAAGAGCACTACGCCACCTTCATTGTAAGGCATTCCAACTTTCAACATGTGAGTT ATTAGGAACTAAATGCCGTCTTTCACAGACAGAACAGGACATTGCCGAGATCGCAGGTAAGGGACTGTCCTTGCA ATCACGAACCTTAACGTAACCTGTCGATTTGGCTTTCAGGCGCTGGTCTAAATTGGATCAGGGTCCCCATTGGAT TTTGGGCGGTTGAGACATGGGAAGGGGAACCATTTTTAGAGAGGACGTCGTGGAAGTAAGTCACTCGTTTCGTCG AACACCGCGGCGATGCTGAATACAATGTCTAGATATTTCCTTCGGATAATCAAATGGGCTAGAAAATACGGCCTG AGAGTGGCCCTGGATCTACATGCAGTCCCTGGATCGCAAAACGGTACGTATCTACTCATCCGTGGATTTCTTTCC CTGAAATGAGAATGCAGGCTACAATCACTCGGGAAGATTATCTCAAGTTAACTTTCTGGCTGGGAACATGGGTAT CGCCAATGCTCAGAGGACGCTCTATACCCTCCGCGTTTTTACTGAATTCATCTCCCAGCCTGAGTATAGAGACGT GATTCAAGTGATTGAGCTCGTAAATGAGCCTTTGGCTGGGGAGATAGGTGCAGAGGCCCTGTCGAGCTTCTACCT GGAAGCGTACAACATGATTAGGAAGATCACAGGAACGGGAGACGGAAATGGGCCAGTAAGCTTCATAATCTCTGC AAGGTCATGGGTCTTTTCTAACGAAGATGCAGTACATCGCCATATCCGACGGTCTGCAACCACTCTCATTGTGGG ATGGCCTCCTTCCTGGGGGTGACCGAGTTATCATGGACGGCCACCCATATTTCGCCTTCGGTGGCATAAACACAG CGCCCATAACCGAACCCTCCGAAGATGGAAAGGCTGGAGGGACGTGGCCGAAAGCTGCGTGTGATAACTGGGGTC CCGCCTTCAACCACAGGTACGTTCCCAATCATAGTGCGTGTACCGCGCTGTCACTCACTGCTATTCCCTCTCAGT CGAAGGACGATTGGCCCCACTGTTGGAGGAGAATTCTCGGCTGCACCCAATGATTGTGGCTTATTCCTACGGGGT GTTGGTATCGAATCACCGCACCCTCAATGTCCGGAGTACGATGACTGGGAGAATTACAATGCCACCATGAAAGAA GGATTGGAAAGCTTCATCATGGCCAGCTTCGACGCGTTGGGAGATTTCTTCTTCTGGACATGGAAGGTCAGTGAC AAGTTTATCCCATTGGCGCATATCTGAGGATTCCCATGTAGATCGGTCCTTCGATCGACGGTCGAGTAGGGACCC CCCTCTGGTCCTACAAGCTAGGACTCGAAAATGGTTGGATACCCAGAGATCCTAGAACCTCGATCGGGAAATGTG TAGCTGTTGGGGGAAATCAAGAGCCTTTCGATGGAGCCTACCCACCATATCGGACGGGGGCTGATCCCTCACCCA CCATTCCAGCCTCGTACTCCTCGCAATACCCTTGGCCACCTGCATCCATCACAGAAGCCGCCGTTCCTGTAGAAC TCCTCCCGACCTACACCAATACAGCACCGATCATTACCCTGCCAGTAGAGTTACCGACTGGGACCCCCACTCCTC TTCCCAGCGGGCTGGACGGCTGGTTCAACGACGACGATGTGGAAGGAGGCCCAGCTGAGATTAATGGATGCCAGT ACCCCGACCGCTATATGGCTGAATTCTCTACCATTCCCACTGCACCTTGTACGGGTCCCACTCCCGCCTAAAACC CTCTCTCGCACACGATGTTGGGGTGAACTCTTACAAGCCGAACTCTTACGGACGACAATTTTAATGAAACAGACC ATATCTATTATATTGATAGTCTACTCCAGCAATCTTACCTACTAGTTGTATGACAATTGTCTCGCTACAATGTGG ATTCAGTTGCGCGAAGGCTCGCATTCCAC |
| Length | 2729 |