CopciAB_435914
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_435914 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 435914 |
| Uniprot id | Functional description | ||
| Location | scaffold_8:2308930..2312104 | Strand | + |
| Gene length (nt) | 3175 | Transcript length (nt) | 2829 |
| CDS length (nt) | 2589 | Protein length (aa) | 862 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Auricularia subglabra | Aurde3_1_1337440 | 22 | 7.393E-7 | 53 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_435914.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 196 | 656 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR040976 | Fungal-type protein kinase |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
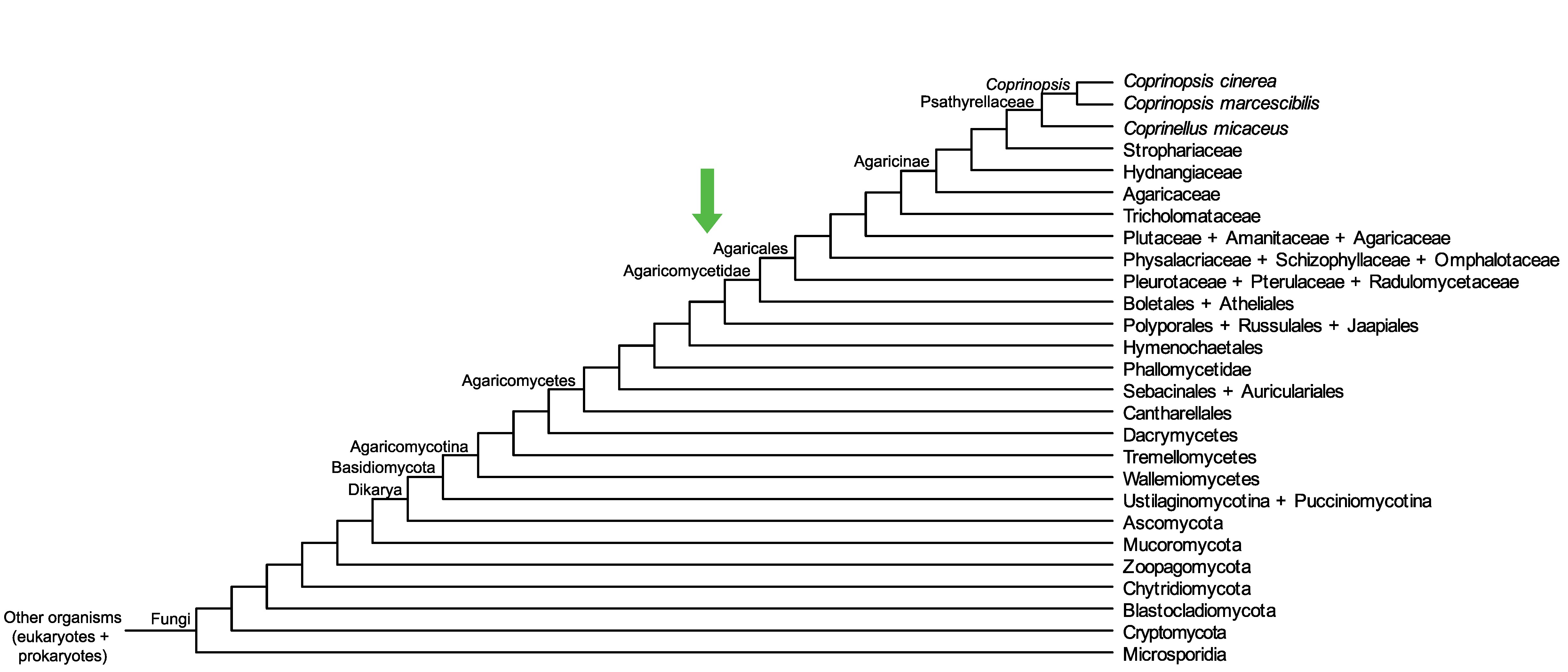
Conservation of CopciAB_435914 across fungi.
Arrow shows the origin of gene family containing CopciAB_435914.
Protein
| Sequence id | CopciAB_435914.T0 |
|---|---|
| Sequence |
>CopciAB_435914.T0 MTLYSLPFLTPPGKKRSTHWSALAKKLVTPRTAPARAGLNATTTQLRGGIAEVMNSEMVGCSIEEFLNHYMPKCN KRTVISVIANLKKENVLVPRAKSKKKKATVSSSEDGRKVYSHVFNNFRPPAIIEESTDANDAEKPNEIMVFDSLA TIGASVIESLRKVKGATVNGHSIRMCPHARLKSPIEGSDHRIDAAMTRNTNLNKRLSITDIAVPFEFKLNRSPGE VYQNRIQLASHVNHAMTDDARRKWTYGITIENDRVSLWYFSRSHSAKSRSFSMVEHPDLFVQILVSLFCATDEQL GFDPLVTLVGDKDYVYEFPPSGERTDSSFYLTLGSLSEFRVLNLVGRNTRIWKVMQVVSAKDHSRVPGTHDMILK DVSLYRNTPTEGDIQRDLFLDIDTFGKTEWRKHELLKDLSEERKAALAQLLEGDGKMYRNYFSCIIDEHVGSPSL PVASQAWDAPDTFQNVEPLEENATETIRKYQATNGLDDDDYEFEDDMSDCGSSVESTDGDDLSRPFEPKQQCRFI YKNVCTPLHNVHTMGEAVDILEQCLLALTLMFCAGWVHRDISTGNILAHRSSPGTPWQVKLSDLEYAKRFPGNPT ATASTEVKTGTMYFMAVEVQSSQLILTDTSEASNTVIIHNPQHDLESLWWILLWLVTMRVRQNLPRAFGKETFQQ TMSGAYIKGRNHCLSYSLTQEQDLLRSLPRPFRSTILEGLEKVRKDLKREYVNRNATNKQTIIGSYSRILGTTFP DFFSGLRARRDEWAPVKLIVSKTVRKEELKRKASKMEGTSDEAKKASTVGKSSGNGRVEPRKYAALPNGHLGQNE TLPGDVSMRPVTRSMTKTTPNTPQNDEKRPFKRSRHC |
| Length | 862 |
Coding
| Sequence id | CopciAB_435914.T0 |
|---|---|
| Sequence |
>CopciAB_435914.T0 ATGACGCTTTACTCGTTGCCGTTTTTAACGCCCCCGGGCAAGAAAAGATCTACCCATTGGAGCGCCCTCGCTAAG AAGCTGGTAACTCCGCGAACCGCACCAGCTAGGGCAGGGCTTAATGCCACAACCACGCAGCTCCGAGGAGGGATC GCGGAAGTGATGAACTCAGAGATGGTCGGCTGCTCGATCGAAGAATTCCTCAATCACTACATGCCCAAGTGCAAC AAGAGGACCGTCATATCGGTCATAGCAAACCTGAAGAAGGAGAACGTCCTCGTACCACGCGCAAAGTCGAAGAAG AAGAAGGCCACAGTATCCTCGAGTGAAGACGGAAGAAAGGTATATTCCCATGTCTTCAACAACTTCCGCCCGCCC GCCATTATCGAGGAGAGTACCGACGCCAACGACGCGGAGAAGCCCAACGAGATAATGGTTTTCGACTCTTTGGCG ACAATCGGTGCATCGGTCATTGAGAGCCTGCGCAAGGTGAAAGGTGCCACTGTCAATGGTCATTCCATTCGCATG TGCCCCCATGCTCGTCTCAAGTCCCCCATCGAAGGGTCGGATCATCGGATTGACGCAGCGATGACAAGAAACACG AATTTGAACAAGCGACTTTCGATCACCGACATTGCAGTGCCCTTTGAATTCAAACTAAATCGTTCGCCGGGCGAG GTTTATCAGAACCGCATTCAGCTCGCGTCTCACGTCAACCACGCCATGACCGATGATGCACGCCGAAAATGGACC TACGGTATTACCATTGAGAACGATCGAGTTTCTCTTTGGTATTTCTCTCGCTCGCATTCGGCCAAATCCAGATCC TTCAGTATGGTCGAGCACCCCGACCTCTTTGTCCAGATTCTGGTTTCGTTATTTTGCGCCACCGACGAGCAACTC GGATTTGACCCACTTGTCACGTTGGTGGGGGATAAAGACTACGTCTACGAGTTCCCGCCCAGTGGCGAGAGGACG GATAGCAGCTTCTATCTCACCCTCGGTTCACTCTCCGAATTTCGTGTTCTAAACCTGGTTGGGCGCAACACCCGC ATCTGGAAGGTTATGCAAGTCGTCTCCGCGAAGGATCATAGTCGGGTCCCGGGTACGCACGACATGATACTCAAG GATGTCTCCCTTTATCGCAACACACCGACGGAGGGGGACATCCAGAGAGATCTATTCCTTGACATCGACACCTTT GGGAAAACGGAATGGCGAAAGCACGAGTTGTTGAAGGACCTATCTGAGGAAAGGAAGGCGGCTTTGGCACAGTTG CTGGAAGGCGACGGAAAGATGTACAGGAACTATTTCTCATGCATTATCGACGAGCACGTTGGGAGCCCTAGCCTT CCTGTCGCAAGTCAGGCATGGGATGCCCCTGACACCTTCCAGAATGTCGAGCCCTTGGAGGAGAATGCTACTGAG ACGATTCGTAAATACCAGGCCACAAATGGTTTGGATGACGATGATTATGAATTTGAAGACGACATGTCAGACTGT GGAAGCAGTGTCGAAAGTACAGATGGAGATGATCTTTCTCGGCCCTTCGAACCGAAACAACAGTGCAGATTCATC TACAAGAACGTATGCACCCCACTTCACAACGTCCACACCATGGGAGAGGCAGTTGATATCCTAGAGCAATGTCTT TTGGCTCTTACCCTGATGTTCTGCGCGGGATGGGTTCACCGGGACATCAGCACCGGAAACATACTCGCCCATCGT TCCTCGCCAGGTACACCATGGCAGGTCAAGTTGTCCGATCTCGAGTACGCCAAACGTTTTCCGGGCAACCCTACA GCAACTGCAAGTACGGAGGTCAAAACGGGAACCATGTACTTCATGGCTGTCGAGGTCCAAAGCTCACAGCTTATC CTCACTGATACTTCCGAGGCAAGCAACACAGTAATCATTCACAACCCCCAGCACGATCTCGAATCTCTATGGTGG ATCTTGCTCTGGCTCGTGACAATGCGAGTGAGGCAGAATCTCCCCCGAGCGTTTGGAAAAGAGACATTCCAACAG ACCATGTCTGGTGCTTATATCAAGGGGCGCAACCACTGTCTGTCGTATTCCCTAACACAGGAGCAAGACCTGCTC CGCTCATTGCCCCGACCTTTCCGGTCAACCATCCTTGAAGGGCTCGAGAAGGTCCGAAAAGATCTGAAGCGGGAG TACGTCAATCGGAATGCCACCAACAAGCAGACCATAATTGGTTCCTATTCACGCATTCTGGGAACAACCTTTCCG GACTTCTTCTCTGGATTGCGTGCCAGGAGGGACGAATGGGCGCCTGTCAAATTGATTGTGTCAAAGACAGTCCGC AAAGAAGAGCTAAAGCGCAAGGCCTCAAAGATGGAAGGGACTAGCGACGAAGCAAAGAAGGCCTCTACTGTGGGT AAGAGCAGCGGCAATGGCAGAGTGGAACCACGGAAATATGCCGCCTTACCGAATGGGCACCTTGGTCAAAACGAG ACCCTCCCCGGCGACGTGTCTATGAGACCCGTAACTAGGTCGATGACCAAGACTACCCCCAATACGCCTCAGAAT GATGAGAAGAGACCTTTTAAGCGATCTCGCCATTGCTGA |
| Length | 2589 |
Transcript
| Sequence id | CopciAB_435914.T0 |
|---|---|
| Sequence |
>CopciAB_435914.T0 CACTACCATCCATTGCGGGAGTTGGCCGTTAACAGAATACGAAGTATACTCTCTATATCCAAAGTACCCAGAGCG CAACGATGACGCTTTACTCGTTGCCGTTTTTAACGCCCCCGGGCAAGAAAAGATCTACCCATTGGAGCGCCCTCG CTAAGAAGCTGGTAACTCCGCGAACCGCACCAGCTAGGGCAGGGCTTAATGCCACAACCACGCAGCTCCGAGGAG GGATCGCGGAAGTGATGAACTCAGAGATGGTCGGCTGCTCGATCGAAGAATTCCTCAATCACTACATGCCCAAGT GCAACAAGAGGACCGTCATATCGGTCATAGCAAACCTGAAGAAGGAGAACGTCCTCGTACCACGCGCAAAGTCGA AGAAGAAGAAGGCCACAGTATCCTCGAGTGAAGACGGAAGAAAGGTATATTCCCATGTCTTCAACAACTTCCGCC CGCCCGCCATTATCGAGGAGAGTACCGACGCCAACGACGCGGAGAAGCCCAACGAGATAATGGTTTTCGACTCTT TGGCGACAATCGGTGCATCGGTCATTGAGAGCCTGCGCAAGGTGAAAGGTGCCACTGTCAATGGTCATTCCATTC GCATGTGCCCCCATGCTCGTCTCAAGTCCCCCATCGAAGGGTCGGATCATCGGATTGACGCAGCGATGACAAGAA ACACGAATTTGAACAAGCGACTTTCGATCACCGACATTGCAGTGCCCTTTGAATTCAAACTAAATCGTTCGCCGG GCGAGGTTTATCAGAACCGCATTCAGCTCGCGTCTCACGTCAACCACGCCATGACCGATGATGCACGCCGAAAAT GGACCTACGGTATTACCATTGAGAACGATCGAGTTTCTCTTTGGTATTTCTCTCGCTCGCATTCGGCCAAATCCA GATCCTTCAGTATGGTCGAGCACCCCGACCTCTTTGTCCAGATTCTGGTTTCGTTATTTTGCGCCACCGACGAGC AACTCGGATTTGACCCACTTGTCACGTTGGTGGGGGATAAAGACTACGTCTACGAGTTCCCGCCCAGTGGCGAGA GGACGGATAGCAGCTTCTATCTCACCCTCGGTTCACTCTCCGAATTTCGTGTTCTAAACCTGGTTGGGCGCAACA CCCGCATCTGGAAGGTTATGCAAGTCGTCTCCGCGAAGGATCATAGTCGGGTCCCGGGTACGCACGACATGATAC TCAAGGATGTCTCCCTTTATCGCAACACACCGACGGAGGGGGACATCCAGAGAGATCTATTCCTTGACATCGACA CCTTTGGGAAAACGGAATGGCGAAAGCACGAGTTGTTGAAGGACCTATCTGAGGAAAGGAAGGCGGCTTTGGCAC AGTTGCTGGAAGGCGACGGAAAGATGTACAGGAACTATTTCTCATGCATTATCGACGAGCACGTTGGGAGCCCTA GCCTTCCTGTCGCAAGTCAGGCATGGGATGCCCCTGACACCTTCCAGAATGTCGAGCCCTTGGAGGAGAATGCTA CTGAGACGATTCGTAAATACCAGGCCACAAATGGTTTGGATGACGATGATTATGAATTTGAAGACGACATGTCAG ACTGTGGAAGCAGTGTCGAAAGTACAGATGGAGATGATCTTTCTCGGCCCTTCGAACCGAAACAACAGTGCAGAT TCATCTACAAGAACGTATGCACCCCACTTCACAACGTCCACACCATGGGAGAGGCAGTTGATATCCTAGAGCAAT GTCTTTTGGCTCTTACCCTGATGTTCTGCGCGGGATGGGTTCACCGGGACATCAGCACCGGAAACATACTCGCCC ATCGTTCCTCGCCAGGTACACCATGGCAGGTCAAGTTGTCCGATCTCGAGTACGCCAAACGTTTTCCGGGCAACC CTACAGCAACTGCAAGTACGGAGGTCAAAACGGGAACCATGTACTTCATGGCTGTCGAGGTCCAAAGCTCACAGC TTATCCTCACTGATACTTCCGAGGCAAGCAACACAGTAATCATTCACAACCCCCAGCACGATCTCGAATCTCTAT GGTGGATCTTGCTCTGGCTCGTGACAATGCGAGTGAGGCAGAATCTCCCCCGAGCGTTTGGAAAAGAGACATTCC AACAGACCATGTCTGGTGCTTATATCAAGGGGCGCAACCACTGTCTGTCGTATTCCCTAACACAGGAGCAAGACC TGCTCCGCTCATTGCCCCGACCTTTCCGGTCAACCATCCTTGAAGGGCTCGAGAAGGTCCGAAAAGATCTGAAGC GGGAGTACGTCAATCGGAATGCCACCAACAAGCAGACCATAATTGGTTCCTATTCACGCATTCTGGGAACAACCT TTCCGGACTTCTTCTCTGGATTGCGTGCCAGGAGGGACGAATGGGCGCCTGTCAAATTGATTGTGTCAAAGACAG TCCGCAAAGAAGAGCTAAAGCGCAAGGCCTCAAAGATGGAAGGGACTAGCGACGAAGCAAAGAAGGCCTCTACTG TGGGTAAGAGCAGCGGCAATGGCAGAGTGGAACCACGGAAATATGCCGCCTTACCGAATGGGCACCTTGGTCAAA ACGAGACCCTCCCCGGCGACGTGTCTATGAGACCCGTAACTAGGTCGATGACCAAGACTACCCCCAATACGCCTC AGAATGATGAGAAGAGACCTTTTAAGCGATCTCGCCATTGCTGAACACGCTGAGTCCTAATGCTGCCAGACTTGT ACATATGTCATTTAGCCCAAGGAAGGTTACAGGGAACTACAGTGTATTTCTCGCAGGTCGACGGGCTTCAGAATT ATTTTTGTGTACACTTATCAACATCAATACATCCATTATCAGTTTCACCCCAAA |
| Length | 2829 |
Gene
| Sequence id | CopciAB_435914.T0 |
|---|---|
| Sequence |
>CopciAB_435914.T0 CACTACCATCCATTGCGGGAGTTGGCCGTTAACAGAATACGAAGTATACTCTCTATATCCAAAGTACCCAGAGCG CAACGATGACGCTTTACTCGTTGCCGTTTTTGTATGTCCAACTTGGCCACTTGACTCTCCTCTTCTCACTGACGC CTCGAATAGAACGCCCCCGGGCAAGAAAAGATCTACCCATTGGAGCGCCCTCGCTAAGAAGCTGGTAACTCCGCG AACCGCACCAGCTAGGGCAGGGCTTAATGCCACAACCACGCAGCTCCGAGGAGGGATCGCGGAAGTGATGAACTC AGAGATGGTCGGCTGCTCGATCGAAGAATTCCTCAATCACTACATGCCCAAGTGCAACAAGAGGACCGTCATATC GGTCATAGCAAACCTGAAGAAGGAGAACGTCCTCGTACCACGCGCAAAGTCGAAGAAGAAGAAGGCCACAGTATC CTCGAGTGAAGACGGAAGAAAGGTATATTCCCATGTCTTCAACAACTTCCGCCCGCCCGCCATTATCGAGGAGAG TACCGACGCCAACGACGCGGAGAAGCCCAACGAGATAATGGTTTTCGACTCTTTGGCGACAATCGGTGCATCGGT CATTGAGAGCCTGCGCAAGGTGAAAGGTGCCACTGTCAATGGTCATTCCATTCGCATGTGCCCCCATGCTCGTCT CAAGTCCCCCATCGAAGGGTCGGATCATCGGATTGACGCAGCGATGACAAGAAACACGAATTTGAACAAGCGACT TTCGATCACCGACATTGCAGTGCCCTTTGAATTCAAACTAAATCGTTCGCCGGGCGAGGTTTATCAGGTATGAGA GCACTGTATCCGGGATTGTCTGAATATACCACTAATGAGCGCGTTCAGAACCGCATTCAGCTCGCGTCTCACGTC AACCACGCCATGACCGATGATGCACGCCGAAAATGGACCTACGGTGTAAGTCCTGTGTTCACACTGCCAGTAGAC CGGTACTCATTGTCCCCTGTTCTAGATTACCATTGAGAACGATCGAGTTTCTCTTTGGTATTTCTCTCGCTCGCA TTCGGCCAAATCCAGATCCTTCAGTATGGTCGAGGTACGTTCAACATTAATTAGTCCTTTGAGATCGTTTCGTAT CTAACCCCGTCTTTCAGCACCCCGACCTCTTTGTCCAGATTCTGGTTTCGTTATTTTGCGCCACCGACGAGCAAC TCGGATTTGACCCACTTGTCACGTTGGTGGGGGATAAAGACTACGTCTACGAGTTCCCGCCCAGTGGCGAGAGGA CGGATAGCAGCTTCTATCTCACCCTCGGTTCACTCTCCGAATTTCGTGTTCTAAACCTGGTTGGGCGCAACACCC GCATCTGGAAGGTTATGCAAGTCGTCTCCGCGAAGGATCATAGTCGGGTCCCGGGTACGCACGACATGATACTCA AGGATGTCTCCCTTTATCGCAACACACCGACGGAGGGGGACATCCAGAGAGATCTATTCCTTGACATCGACACCT TTGGGAAAACGGAATGGCGAAAGCACGAGTTGTTGAAGGACCTATCTGAGGAAAGGAAGGCGGCTTTGGCACAGT TGCTGGAAGGCGACGGAAAGATGTACAGGAACTATTTCTCATGCATTATCGACGAGCACGTTGGGAGCCCTAGCC TTCCTGTCGCAAGTCAGGCATGGGATGCCCCTGACACCTTCCAGAATGTCGAGCCCTTGGAGGAGAATGCTACTG AGACGATTCGTAAATACCAGGCCACAAATGGTTTGGATGACGATGATTATGAATTTGAAGACGACATGTCAGACT GTGGAAGCAGTGTCGAAAGTACAGATGGAGATGATCTTTCTCGGCCCTTCGAACCGAAACAACAGTGCAGATTCA TCTACAAGAACGTATGCACCCCACTTCACAACGTCCACACCATGGGAGAGGCAGTTGATATCCTAGAGCAATGTC TTTTGGGTAAGTGGCGTCCAAGCTTTCTGAGGCTTGCTGTAGCGGCCACTGATTATCATTCGATTACAGCTCTTA CCCTGATGTTCTGCGCGGGATGGGTTCACCGGGACATCAGCACCGGAAACATACTCGCCCATCGTTCCTCGCCAG GTACACCATGGCAGGTCAAGTTGTCCGATCTCGAGTACGCCAAACGTTTTCCGGGCAACCCTACAGCAACTGCAA GTACGGAGGTCAAAACGGTAATGTCCCACGATCTTGGTCACATAGTCTCAATTGAAACTCACTCGCTTCACATTT CAGGGAACCATGTACTTCATGGCTGTCGAGGTCCAAAGCTCACAGCTTATCCTCACTGATACTTCCGAGGCAAGC AACACAGTAATCATTCACAACCCCCAGCACGATCTCGAATCTCTATGGTGGATCTTGCTCTGGCTCGTGACAATG CGAGTGAGGCAGAATCTCCCCCGAGCGTTTGGAAAAGAGACATTCCAACAGACCATGTCTGGTGCTTATATCAAG GGGCGCAACCACTGTCTGTCGTATTCCCTAACACAGGAGCAAGACCTGCTCCGCTCATTGCCCCGACCTTTCCGG TCAACCATCCTTGAAGGGCTCGAGAAGGTCCGAAAAGATCTGAAGCGGGAGTACGTCAATCGGAATGCCACCAAC AAGCAGACCATAATTGGTTCCTATTCACGCATTCTGGGAACAACCTTTCCGGACTTCTTCTCTGGATTGCGTGCC AGGAGGGACGAATGGGCGCCTGTCAAATTGATTGTGTCAAAGACAGTCCGCAAAGAAGAGCTAAAGCGCAAGGCC TCAAAGATGGAAGGGACTAGCGACGAAGCAAAGAAGGCCTCTACTGTGGGTAAGAGCAGCGGCAATGGCAGAGTG GAACCACGGAAATATGCCGCCTTACCGAATGGGCACCTTGGTCAAAACGAGACCCTCCCCGGCGACGTGTCTATG AGACCCGTAACTAGGTCGATGACCAAGACTACCCCCAATACGCCTCAGAATGATGAGAAGAGACCTTTTAAGCGA TCTCGCCATTGCTGAACACGCTGAGTCCTAATGCTGCCAGACTTGTACATATGTCATTTAGCCCAAGGAAGGTTA CAGGGAACTACAGTGTATTTCTCGCAGGTCGACGGGCTTCAGAATTATTTTTGTGTACACTTATCAACATCAATA CATCCATTATCAGTTTCACCCCAAA |
| Length | 3175 |