CopciAB_438455
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_438455 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 438455 |
| Uniprot id | Functional description | other AgaK1 protein kinase | |
| Location | scaffold_11:880182..881931 | Strand | + |
| Gene length (nt) | 1750 | Transcript length (nt) | 1408 |
| CDS length (nt) | 1158 | Protein length (aa) | 385 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7266294 | 29.9 | 6.392E-29 | 120 |
| Schizophyllum commune H4-8 | Schco3_2509770 | 30.9 | 2.638E-24 | 106 |
| Lentinula edodes NBRC 111202 | Lenedo1_1061534 | 29.8 | 3.361E-19 | 90 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_21584 | 28.5 | 3.027E-18 | 87 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_438455.T0 |
| Description | other AgaK1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 192 | 229 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | other AgaK1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
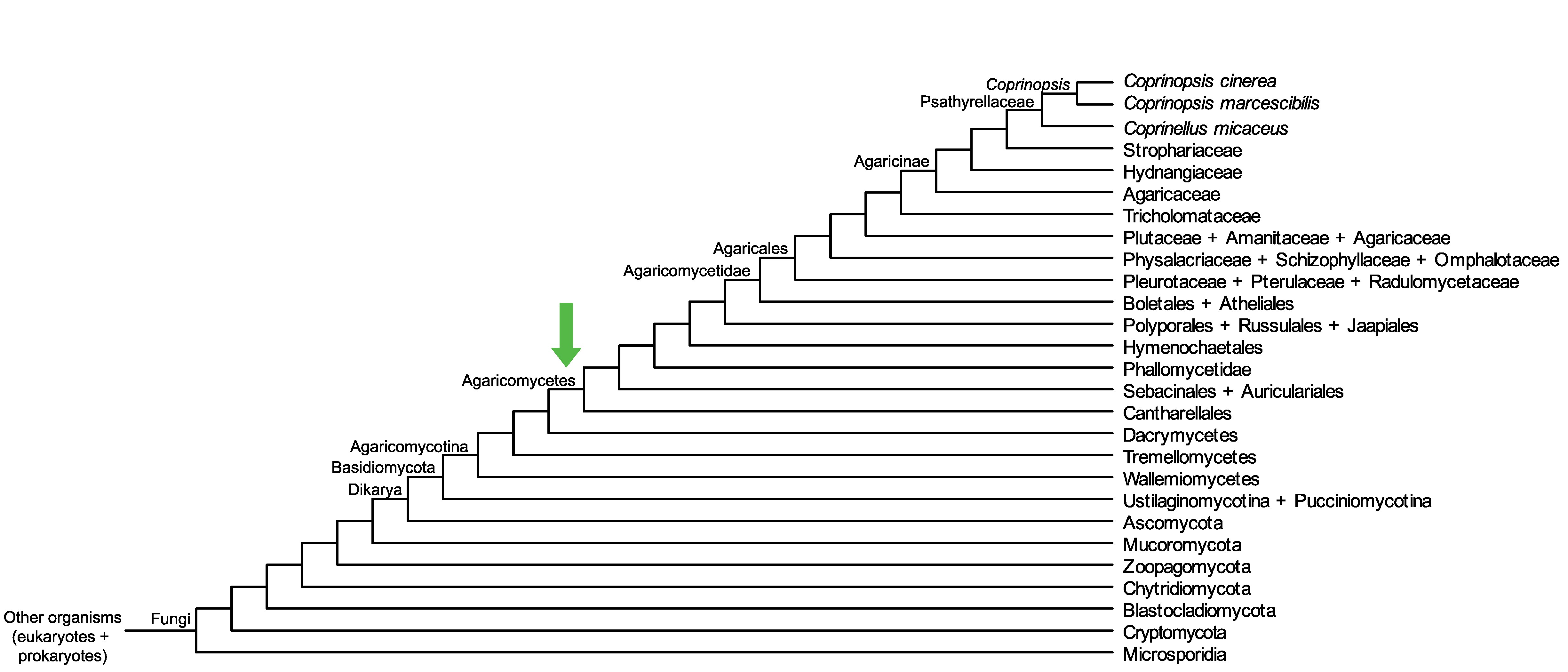
Conservation of CopciAB_438455 across fungi.
Arrow shows the origin of gene family containing CopciAB_438455.
Protein
| Sequence id | CopciAB_438455.T0 |
|---|---|
| Sequence |
>CopciAB_438455.T0 MKLVRCGSLWETLRWTLHAWWHGYHVPTDRVTWTSTRDTTIPYEEEVKITAKRWSLLVPFFASRGYHLYESIIGS RTMPPSSVKPQRGMDDCYPYARRRERHERGLEFIEIQSCRVWAARDNAGREVVIKLVSLGEQEWDELKIYKRLNT PEARADPRNRTLPVLDYIIYNGLVFVVTPRWGEPVVGLSFSKVEQVLVMADQLLEGMAFLHHQRIAHRDVDTRNV VMNALYEIAEGHDCDEIRDSSVVRYAYIDFEASVILPPDTDIGKVVMKREERLTTIRAGLESGESNPFADDIYVL IVVLQRWVRVVENTVPAIGKFFDEIISNYSSDLSSIEILSKFRRIRAGLTPEQLKSPTPGRFWHKGKVQRHLSDW GFAGSAPAST |
| Length | 385 |
Coding
| Sequence id | CopciAB_438455.T0 |
|---|---|
| Sequence |
>CopciAB_438455.T0 ATGAAGTTGGTACGCTGTGGAAGCCTGTGGGAAACTCTCCGGTGGACGCTGCATGCTTGGTGGCATGGTTATCAT GTCCCAACAGATAGGGTTACGTGGACCTCGACGAGGGACACAACTATACCTTATGAAGAGGAGGTCAAGATAACT GCTAAGCGCTGGTCACTACTGGTGCCATTCTTTGCCTCCCGTGGCTATCACCTCTATGAATCGATCATTGGGTCC CGGACCATGCCACCATCCTCAGTCAAACCACAGAGGGGCATGGACGACTGCTATCCCTATGCACGACGCCGAGAG AGGCATGAGAGAGGTCTCGAGTTCATTGAAATACAGAGTTGTAGGGTGTGGGCCGCGCGTGACAATGCCGGAAGA GAGGTCGTCATCAAGCTGGTCTCCTTGGGGGAGCAAGAGTGGGACGAGCTCAAGATTTACAAGAGGTTGAATACC CCTGAAGCTAGGGCAGATCCTCGCAACCGGACCCTGCCTGTCCTCGACTACATTATCTACAATGGGCTGGTATTC GTTGTCACACCCCGCTGGGGGGAGCCAGTCGTGGGGTTGAGCTTCTCTAAGGTAGAGCAGGTCCTTGTCATGGCT GATCAGCTGCTGGAGGGCATGGCATTTCTGCACCACCAACGAATAGCCCACCGCGACGTTGACACGCGGAACGTG GTCATGAACGCGCTATACGAGATTGCCGAGGGACACGACTGCGACGAAATTCGTGACTCTTCCGTTGTGCGTTAC GCTTACATTGACTTTGAAGCTTCGGTAATTCTGCCTCCGGATACGGACATCGGGAAAGTTGTTATGAAGCGGGAG GAGAGGCTCACAACCATACGCGCTGGTCTGGAAAGTGGAGAATCGAATCCCTTTGCAGACGACATCTACGTTTTG ATTGTTGTCTTGCAACGTTGGGTTCGGGTTGTCGAGAATACCGTTCCAGCAATCGGGAAGTTCTTCGACGAGATA ATATCAAATTATTCCAGCGATCTCTCATCCATAGAGATTCTGAGCAAATTTCGCCGCATCCGGGCTGGACTCACA CCAGAACAGTTGAAGAGTCCCACCCCTGGCAGGTTTTGGCATAAAGGAAAGGTGCAAAGACATCTCAGCGACTGG GGTTTTGCGGGGAGTGCACCCGCAAGTACTTAA |
| Length | 1158 |
Transcript
| Sequence id | CopciAB_438455.T0 |
|---|---|
| Sequence |
>CopciAB_438455.T0 AGGAACATTGACAGCTGCCCCCTTGACCTGCTTCTGTAGGCTCTCTCGAACAACACACCACGCTGACGTACCATG AAGTTGGTACGCTGTGGAAGCCTGTGGGAAACTCTCCGGTGGACGCTGCATGCTTGGTGGCATGGTTATCATGTC CCAACAGATAGGGTTACGTGGACCTCGACGAGGGACACAACTATACCTTATGAAGAGGAGGTCAAGATAACTGCT AAGCGCTGGTCACTACTGGTGCCATTCTTTGCCTCCCGTGGCTATCACCTCTATGAATCGATCATTGGGTCCCGG ACCATGCCACCATCCTCAGTCAAACCACAGAGGGGCATGGACGACTGCTATCCCTATGCACGACGCCGAGAGAGG CATGAGAGAGGTCTCGAGTTCATTGAAATACAGAGTTGTAGGGTGTGGGCCGCGCGTGACAATGCCGGAAGAGAG GTCGTCATCAAGCTGGTCTCCTTGGGGGAGCAAGAGTGGGACGAGCTCAAGATTTACAAGAGGTTGAATACCCCT GAAGCTAGGGCAGATCCTCGCAACCGGACCCTGCCTGTCCTCGACTACATTATCTACAATGGGCTGGTATTCGTT GTCACACCCCGCTGGGGGGAGCCAGTCGTGGGGTTGAGCTTCTCTAAGGTAGAGCAGGTCCTTGTCATGGCTGAT CAGCTGCTGGAGGGCATGGCATTTCTGCACCACCAACGAATAGCCCACCGCGACGTTGACACGCGGAACGTGGTC ATGAACGCGCTATACGAGATTGCCGAGGGACACGACTGCGACGAAATTCGTGACTCTTCCGTTGTGCGTTACGCT TACATTGACTTTGAAGCTTCGGTAATTCTGCCTCCGGATACGGACATCGGGAAAGTTGTTATGAAGCGGGAGGAG AGGCTCACAACCATACGCGCTGGTCTGGAAAGTGGAGAATCGAATCCCTTTGCAGACGACATCTACGTTTTGATT GTTGTCTTGCAACGTTGGGTTCGGGTTGTCGAGAATACCGTTCCAGCAATCGGGAAGTTCTTCGACGAGATAATA TCAAATTATTCCAGCGATCTCTCATCCATAGAGATTCTGAGCAAATTTCGCCGCATCCGGGCTGGACTCACACCA GAACAGTTGAAGAGTCCCACCCCTGGCAGGTTTTGGCATAAAGGAAAGGTGCAAAGACATCTCAGCGACTGGGGT TTTGCGGGGAGTGCACCCGCAAGTACTTAATTCATATCTTCTGAGCAGTCTAGCAATCGCCATGACTCTTCCCTG GGGACTTGTGCTTATACAAGGCTGGTGTACATTCGCCAACCACAGGAGTTGGACAGGAGTCGAAAACACCAACGC AGGTCGGATGCTCCCGGCCACAAGCTACGCATGAATGAATACACGAATTGCGTGTGGG |
| Length | 1408 |
Gene
| Sequence id | CopciAB_438455.T0 |
|---|---|
| Sequence |
>CopciAB_438455.T0 AGGAACATTGACAGCTGCCCCCTTGACCTGCTTCTGTAGGCTCTCTCGAACAACACACCACGCTGACGTACCATG AAGTTGGTACGCTGTGGAAGCCTGTGGGAAACTCTCCGGTGGACGCTGCATGCTTGGTGGCATGGTTATCATGTC CCAACAGATAGGGTTACGTGGACCTCGACGAGGGACACAACTATACCTTATGAAGAGGAGGTCAAGATAACTGCT AAGCGCTGGTCACTACTGGTGCCATTCTTTGCCTCCCGTGGCTATCACCTCTATGAATCGATCATTGGGTCCCGG ACCATGCCACCATCCTCAGTCAAACCACAGAGGGGCATGGACGACTGCTATCCCTATGCACGACGCCGAGAGAGG CATGAGAGAGGTCTCGAGTTCATTGAAATACAGGTGCGCCGGAAGCTGCCCTATTTACACGGTCATTTAAATCTG ACTCTTGGATCGTAGAGTTGTAGGGTGTGGGCCGCGCGTGACAATGCCGGAAGAGAGGTCGTCATCAAGTCTCTG TCCCTGTTTATTGGATACAATGACAGAATTAACTGACTTTCACCCCCTTTAGGCTGGTCTCCTTGGGGGAGCAAG AGTGGGACGAGCTCAAGATTTACAAGAGGTTGAATACCCCTGAAGCTAGGGCAGATCCTCGCAACCGGACCCTGC CTGTCCTCGACTACATTATCTACAATGGGCTGGTATTCGTTGTCACACCCCGGTTGGTGTCTCAATACCCCAGCT TCTCTGGCACTGATTTCTCTCATTTATAGCTGGGGGGAGCCAGTCGTGGGGTTGAGCTTCTCTAAGGTAGAGCAG GTCCTTGTCATGGCTGATCAGCTGCTGGAGGTATCACCCACCGCACTACTACTTCGTAATCTGGCTAATTTACCG ATGATCAAGGGCATGGCATTTCTGCACCACCAACGAATAGCCCACCGCGACGTTGACACGCGGAACGTGGTCATG AACGCGCTATACGAGATTGCCGAGGGACACGACTGCGACGAAATTCGTGACTCTTCCGTTGTGCGTTACGCTTAC ATTGACTTTGAAGCTTCGGTAATTCTGCCTCCGGATACGGACATCGGGAAAGTTGTTATGAAGCGGGAGGAGAGG CTCACAACCATACGCGCTGGTCTGGAAAGTGGAGAATCGAATCCCTTTGCAGACGACATCTACGTTTTGATTGTT GTCTTGCAACGTTGGGTTCGGGTGCGTCGTCTGGTAGTCGCTATTTCCGGGAACAGATGGTGCTCACAAACGACT TTCCCCACCAGGTTGTCGAGAATACCGTTCCAGCAATCGGGAAGTTCTTCGACGAGATAATATCAAATTATTCCA GCGATCTCTCATCCATAGAGATTCTGAGCAAATTTCGCCGCATCCGGGCTGGACTCACACCAGAACAGTTGAAGA GTCCCACCCCTGGCAGGTTTTGGCATAAAGGTAATATTAATATCTTCACCTTTTTCGGTATGACCGACGCTGACA CGCGATGTAGGAAAGGTGCAAAGACATCTCAGCGACTGGGGTTTTGCGGGGAGTGCACCCGCAAGTACTTAATTC ATATCTTCTGAGCAGTCTAGCAATCGCCATGACTCTTCCCTGGGGACTTGTGCTTATACAAGGCTGGTGTACATT CGCCAACCACAGGAGTTGGACAGGAGTCGAAAACACCAACGCAGGTCGGATGCTCCCGGCCACAAGCTACGCATG AATGAATACACGAATTGCGTGTGGG |
| Length | 1750 |