CopciAB_441916
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_441916 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 441916 |
| Uniprot id | Functional description | RNA polymerase II CTD heptapeptide repeat kinase activity | |
| Location | scaffold_1:987375..989732 | Strand | - |
| Gene length (nt) | 2358 | Transcript length (nt) | 1861 |
| CDS length (nt) | 1518 | Protein length (aa) | 505 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264903 | 52.3 | 2.628E-168 | 533 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB19371 | 49.5 | 1.847E-134 | 435 |
| Schizophyllum commune H4-8 | Schco3_2593755 | 45.8 | 8.201E-113 | 372 |
| Pleurotus ostreatus PC9 | PleosPC9_1_100969 | 41.1 | 6.949E-111 | 366 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1458455 | 40.9 | 1.969E-109 | 362 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1075570 | 40.9 | 1.674E-109 | 362 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_196486 | 40.4 | 6.077E-99 | 331 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_67661 | 40 | 6.734E-98 | 328 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_148505 | 39 | 1.21E-90 | 307 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_6409 | 38.3 | 1.383E-89 | 304 |
| Lentinula edodes NBRC 111202 | Lenedo1_1090918 | 37.1 | 7.482E-87 | 296 |
| Flammulina velutipes | Flave_chr09AA00012 | 42.6 | 5.597E-75 | 261 |
| Grifola frondosa | Grifr_OBZ79686 | 42 | 3.803E-63 | 226 |
| Lentinula edodes B17 | Lened_B_1_1_14387 | 41.3 | 1.332E-39 | 155 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_441916.T0 |
| Description | RNA polymerase II CTD heptapeptide repeat kinase activity |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 37 | 258 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K01783 |
| K02208 |
EggNOG
| COG category | Description |
|---|---|
| F | RNA polymerase II CTD heptapeptide repeat kinase activity |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
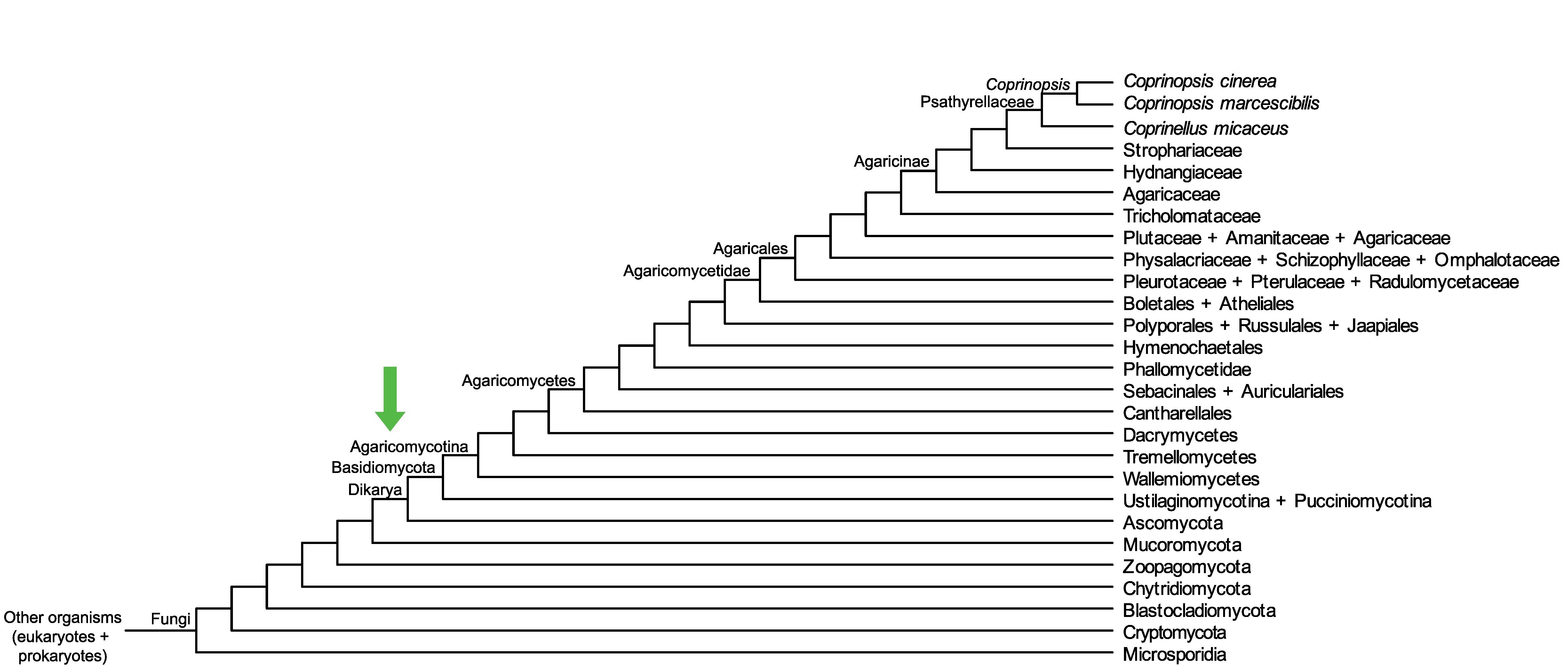
Conservation of CopciAB_441916 across fungi.
Arrow shows the origin of gene family containing CopciAB_441916.
Protein
| Sequence id | CopciAB_441916.T0 |
|---|---|
| Sequence |
>CopciAB_441916.T0 MTLTKADVMRIQATPPSHYDNIKEGPVSSVARAWLSMKNGEPQWIVIKSAPVIRKFSREPHDIRKELRILTELSH RNIISVLGSFDDEETDTLKVYMPYIPISLADLLSSPYFSPHPFPPQLPQDLVALRFHEAQFTTISRSIIVQVLCA VAYLHDESRRIAHRDIKPENILLTKDGCVKLIDFGVCWKEKEPGYSKQYDLWPEYSGKLYFEVCTGAYRAPELLF GTRAYDAIAVDLWSLGATLAEFFTPLRMSSDDTDDDGDDDDGGHCHDDDDDSTEPGNESNGASSDGEYGSSTLQP FIVPRYLRIGYPDAQWHRDTLFNGDRGEIGLAWSIFKIYGTPTLDNWPEFDDLPGAKSVVFNVVPAVPVEPLLPN LPPSTRSSSPLASPASHCNGVLSTPKAYRHPALDPIRKSTPFDLLSRFLVYPSGSRQTASEALRHPWITADSAIL LPKGYEFEMGAGAGVDRDNVAQLKRMMAHEWKGKSLGELLHGVLLAPPGSANPRW |
| Length | 505 |
Coding
| Sequence id | CopciAB_441916.T0 |
|---|---|
| Sequence |
>CopciAB_441916.T0 ATGACCTTAACAAAGGCGGATGTGATGAGGATTCAGGCTACACCTCCATCTCATTACGATAATATCAAAGAAGGC CCAGTTTCATCTGTCGCAAGAGCTTGGTTGTCGATGAAGAATGGGGAACCTCAGTGGATAGTCATCAAGTCGGCC CCTGTGATTCGCAAGTTTTCCAGGGAGCCTCATGACATTCGCAAAGAACTGAGGATCTTGACGGAGCTCTCCCAT CGGAATATCATCTCTGTGTTGGGTAGCTTCGATGACGAGGAAACCGACACGCTCAAAGTGTACATGCCCTATATT CCGATCTCCTTGGCCGACCTCCTCTCTTCCCCATACTTCTCCCCGCACCCATTCCCGCCTCAATTGCCACAGGAT CTGGTTGCTTTGCGATTCCACGAAGCACAGTTCACCACGATATCCCGCTCAATCATCGTTCAAGTGCTGTGCGCC GTGGCTTATCTCCACGACGAATCTCGTCGAATCGCTCACCGTGACATAAAGCCCGAAAACATCCTCCTCACCAAG GATGGTTGTGTCAAGTTGATTGATTTCGGCGTGTGTTGGAAGGAGAAGGAACCAGGGTATTCGAAGCAATATGAC CTCTGGCCAGAATACAGCGGAAAACTTTATTTCGAAGTATGCACTGGAGCCTACCGTGCTCCCGAGCTTCTGTTC GGAACTCGAGCATACGATGCTATTGCGGTTGATCTATGGAGCTTAGGTGCCACCTTAGCAGAGTTCTTCACCCCC CTCCGGATGTCCAGTGATGACACTGATGATGACGGAGACGACGACGATGGTGGGCACTGCCACGACGACGACGAT GACTCTACTGAACCCGGTAACGAGTCAAATGGGGCATCCAGTGACGGAGAATATGGATCATCGACACTCCAGCCC TTCATCGTTCCCCGGTACCTGCGTATAGGTTACCCAGACGCGCAATGGCACCGGGATACCTTGTTCAACGGGGAT CGTGGCGAGATTGGTCTCGCGTGGAGCATCTTCAAGATTTACGGTACGCCCACCCTCGACAACTGGCCGGAATTC GATGACCTTCCAGGAGCGAAGAGCGTCGTCTTCAACGTGGTCCCAGCCGTCCCAGTTGAGCCTCTCCTCCCCAAC TTGCCTCCTTCCACACGTTCTTCATCGCCACTTGCCTCCCCTGCTTCTCACTGCAATGGCGTTTTGAGCACGCCC AAGGCCTATCGCCATCCGGCTCTCGATCCCATTCGTAAATCCACGCCTTTTGATTTATTGTCGAGGTTCCTCGTC TACCCATCTGGCTCCCGCCAGACAGCTTCTGAAGCTCTCCGGCATCCTTGGATTACTGCCGACTCGGCGATCCTC TTGCCTAAAGGATACGAGTTCGAAATGGGCGCTGGAGCGGGAGTTGATCGGGACAACGTCGCGCAGTTGAAGAGG ATGATGGCTCATGAATGGAAGGGCAAGTCTTTGGGAGAGCTGTTGCATGGAGTGCTGTTGGCTCCTCCTGGTTCG GCGAATCCGCGATGGTAA |
| Length | 1518 |
Transcript
| Sequence id | CopciAB_441916.T0 |
|---|---|
| Sequence |
>CopciAB_441916.T0 AGTCTCTTCGACCGTCACAACCAACCCGCTGCGAATGGCGCTCTGCTATTCACAAGTGGCGGGATAGTTCGCCAT TTACCTGTTCGACTTCGACAAACGTTTTCTACCACTACAACGGGGAAAGGCTGCCACCATGACCTTAACAAAGGC GGATGTGATGAGGATTCAGGCTACACCTCCATCTCATTACGATAATATCAAAGAAGGCCCAGTTTCATCTGTCGC AAGAGCTTGGTTGTCGATGAAGAATGGGGAACCTCAGTGGATAGTCATCAAGTCGGCCCCTGTGATTCGCAAGTT TTCCAGGGAGCCTCATGACATTCGCAAAGAACTGAGGATCTTGACGGAGCTCTCCCATCGGAATATCATCTCTGT GTTGGGTAGCTTCGATGACGAGGAAACCGACACGCTCAAAGTGTACATGCCCTATATTCCGATCTCCTTGGCCGA CCTCCTCTCTTCCCCATACTTCTCCCCGCACCCATTCCCGCCTCAATTGCCACAGGATCTGGTTGCTTTGCGATT CCACGAAGCACAGTTCACCACGATATCCCGCTCAATCATCGTTCAAGTGCTGTGCGCCGTGGCTTATCTCCACGA CGAATCTCGTCGAATCGCTCACCGTGACATAAAGCCCGAAAACATCCTCCTCACCAAGGATGGTTGTGTCAAGTT GATTGATTTCGGCGTGTGTTGGAAGGAGAAGGAACCAGGGTATTCGAAGCAATATGACCTCTGGCCAGAATACAG CGGAAAACTTTATTTCGAAGTATGCACTGGAGCCTACCGTGCTCCCGAGCTTCTGTTCGGAACTCGAGCATACGA TGCTATTGCGGTTGATCTATGGAGCTTAGGTGCCACCTTAGCAGAGTTCTTCACCCCCCTCCGGATGTCCAGTGA TGACACTGATGATGACGGAGACGACGACGATGGTGGGCACTGCCACGACGACGACGATGACTCTACTGAACCCGG TAACGAGTCAAATGGGGCATCCAGTGACGGAGAATATGGATCATCGACACTCCAGCCCTTCATCGTTCCCCGGTA CCTGCGTATAGGTTACCCAGACGCGCAATGGCACCGGGATACCTTGTTCAACGGGGATCGTGGCGAGATTGGTCT CGCGTGGAGCATCTTCAAGATTTACGGTACGCCCACCCTCGACAACTGGCCGGAATTCGATGACCTTCCAGGAGC GAAGAGCGTCGTCTTCAACGTGGTCCCAGCCGTCCCAGTTGAGCCTCTCCTCCCCAACTTGCCTCCTTCCACACG TTCTTCATCGCCACTTGCCTCCCCTGCTTCTCACTGCAATGGCGTTTTGAGCACGCCCAAGGCCTATCGCCATCC GGCTCTCGATCCCATTCGTAAATCCACGCCTTTTGATTTATTGTCGAGGTTCCTCGTCTACCCATCTGGCTCCCG CCAGACAGCTTCTGAAGCTCTCCGGCATCCTTGGATTACTGCCGACTCGGCGATCCTCTTGCCTAAAGGATACGA GTTCGAAATGGGCGCTGGAGCGGGAGTTGATCGGGACAACGTCGCGCAGTTGAAGAGGATGATGGCTCATGAATG GAAGGGCAAGTCTTTGGGAGAGCTGTTGCATGGAGTGCTGTTGGCTCCTCCTGGTTCGGCGAATCCGCGATGGTA ACAATCCCAGCTCGATAGCAGTATAAGCTCTTTAGTGAAACTCTCTCTCTAATTATACTTACCGACCATCGTGTC AGGTTTGGACTGCCTCCCGGTTATGGGATACTGTCTTGACCTTGTTCATTTCTTTCATATTACTTTATGCCGTGT TTTCGATTGTAATTTGAGGAGGGGAAGAAGTTTGAATACCTAATGTAACTTTATCTACGCG |
| Length | 1861 |
Gene
| Sequence id | CopciAB_441916.T0 |
|---|---|
| Sequence |
>CopciAB_441916.T0 AGTCTCTTCGACCGTCACAACCAACCCGCTGCGAATGGCGCTCTGCTATTCACAAGTGGCGGGATAGTTCGCCAT TTACCTGTTCGACTTCGACAAACGTTTTCTACCACTACAACGGGGAAAGGCTGCCACCATGACCTTAACAAAGGC GGATGTGATGAGGATTCAGGCTACAGTGAGTCGAGTGACCATCTATGACCGATTCTTGCCAGCCGCTGGCGACTA GAACAGGCTGCTAATACAGTCTATATTGTCTCTTTGCCTTGATTTTACGCTCCCGGCGGCCCAACTCGATAACTG ACTGGCTTTTTCGCGTCGTTTAGCCTCCATCTCATTACGATAATATCAAAGAAGGCCCAGTTTCATCTGTCGCAA GAGCTTGGTTGTCGATGAAGAATGGGGAACCTCAGTGGATAGTCATCAAGTCGGCCCCTGTGATTCGCAAGTTTT CCAGGGAGCCTCATGACATTCGCAAAGAACTGAGGATCTTGACGGAGCTCTCCCATCGGAATGTGAGTATCTAGC CTCGCGCGCACATGATGCCTTGCTTTTGTGGTCGTATGACCATCGTAATATGGGATCGTCAAGCAAAACCTTGAT CGCTAGTCGTGCATAGATTTTCCTAGCACGGCAGCGATGCGGGTGCACGACCAATTTTTGGGCATGCGATTTATG TCTAAGTAGTTCTGACGATGGCTTTCCAGATCATCTCTGTGTTGGGTAGCTTCGATGACGAGGAAACCGACACGC TCAAAGTGTACATGCCCTATATTCCGATCTCCTTGGCCGACCTCCTCTCTTCCCCATACTTCTCCCCGCACCCAT TCCCGCCTCAATTGCCACAGGATCTGGTTGCTTTGCGATTCCACGAAGCACAGTTCACCACGATATCCCGCTCAA TCATCGTTCAAGTGCTGTGCGCCGTGGCTTATCTCCACGACGAATCTCGTCGAATCGCTCACCGTGACATAAAGC CCGAAAACATCCTCCTCACCAAGGATGGTTGTGTCAAGTTGATTGATTTCGGCGTGTGTTGGAAGGAGAAGGAAC CAGGGTATTCGAAGCAATATGACCTCTGGCCAGAATACAGCGGAAAACTTTATTTCGAAGTATGCACTGGGTGAG TCAGACAAAGTTTACGCAGCTCCCCCATATTATTCTTCTCCATTGGAATGCACCACGGGGCTAACTCAAACCATG CGTATTTTCCCTAATTTAGAGCCTACCGTGCTCCCGAGCTTCTGTTCGGAACTCGAGCATACGATGCTATTGCGG TTGATCTATGGAGCTTAGGTGCCACCTTAGCAGAGTTCTTCACCCCCCTCCGGATGTCCAGTGATGACACTGATG ATGACGGAGACGACGACGATGGTGGGCACTGCCACGACGACGACGATGACTCTACTGAACCCGGTAACGAGTCAA ATGGGGCATCCAGTGACGGAGAATATGGATCATCGACACTCCAGCCCTTCATCGTTCCCCGGTACCTGCGTATAG GTTACCCAGACGCGCAATGGCACCGGGATACCTTGTTCAACGGGGATCGTGGCGAGATTGGTCTCGCGTGGAGCA TCTTCAAGATTTACGGTACGCCCACCCTCGACAACTGGCCGGTATGTCTATGCAGTTTCTCTCGCGTGAGGACAC TAGCCAAATGATGTCCTTTTCCAGGAATTCGATGACCTTCCAGGAGCGAAGAGCGTCGTCTTCAACGTGGTCCCA GCCGTCCCAGTTGAGCCTCTCCTCCCCAACTTGCCTCCTTCCACACGTTCTTCATCGCCACTTGCCTCCCCTGCT TCTCACTGCAATGGCGTTTTGAGCACGCCCAAGGCCTATCGCCATCCGGCTCTCGATCCCATTCGTAAATCCACG CCTTTTGATTTATTGTCGAGGTTCCTCGTCTACCCATCTGGCTCCCGCCAGACAGCTTCTGAAGCTCTCCGGCAT CCTTGGATTACTGCCGACTCGGCGATCCTCTTGCCTAAAGGATACGAGTTCGAAATGGGCGCTGGAGCGGGAGTT GATCGGGACAACGTCGCGCAGTTGAAGAGGATGATGGCTCATGAATGGAAGGGCAAGTCTTTGGGAGAGCTGTTG CATGGAGTGCTGTTGGCTCCTCCTGGTTCGGCGAATCCGCGATGGTAACAATCCCAGCTCGATAGCAGTATAAGC TCTTTAGTGAAACTCTCTCTCTAATTATACTTACCGACCATCGTGTCAGGTTTGGACTGCCTCCCGGTTATGGGA TACTGTCTTGACCTTGTTCATTTCTTTCATATTACTTTATGCCGTGTTTTCGATTGTAATTTGAGGAGGGGAAGA AGTTTGAATACCTAATGTAACTTTATCTACGCG |
| Length | 2358 |