CopciAB_443321
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_443321 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | YPK2 | Synonyms | 443321 |
| Uniprot id | Functional description | Haspin like kinase domain | |
| Location | scaffold_7:1675406..1678143 | Strand | + |
| Gene length (nt) | 2738 | Transcript length (nt) | 2162 |
| CDS length (nt) | 1662 | Protein length (aa) | 553 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7271336 | 86.8 | 0 | 1008 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB18028 | 83.8 | 1.848E-304 | 964 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_193872 | 82 | 4.428E-304 | 945 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114034 | 82.1 | 4.615E-304 | 945 |
| Lentinula edodes NBRC 111202 | Lenedo1_1039497 | 81.5 | 3.53E-304 | 937 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_2296 | 81.5 | 3.503E-304 | 937 |
| Flammulina velutipes | Flave_chr05AA00262 | 79 | 3.291E-291 | 911 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_116859 | 77 | 3.296E-289 | 885 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1442416 | 77.9 | 1.783E-288 | 883 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1094051 | 79.2 | 4.396E-285 | 873 |
| Pleurotus ostreatus PC9 | PleosPC9_1_116440 | 79.2 | 4.303E-285 | 873 |
| Schizophyllum commune H4-8 | Schco3_2615950 | 76.5 | 6.365E-284 | 870 |
| Grifola frondosa | Grifr_OBZ71365 | 75.8 | 3.308E-280 | 859 |
| Auricularia subglabra | Aurde3_1_1271841 | 62.6 | 1.464E-223 | 696 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | YPK2 |
|---|---|
| Protein id | CopciAB_443321.T0 |
| Description | Haspin like kinase domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd11651 | YPK1_N_like | - | 40 | 220 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 226 | 480 |
| Pfam | PF00433 | Protein kinase C terminal domain | IPR017892 | 504 | 543 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017892 | Protein kinase, C-terminal |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
| IPR000719 | Protein kinase domain |
| IPR000961 | AGC-kinase, C-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| K13303 |
EggNOG
| COG category | Description |
|---|---|
| T | Haspin like kinase domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
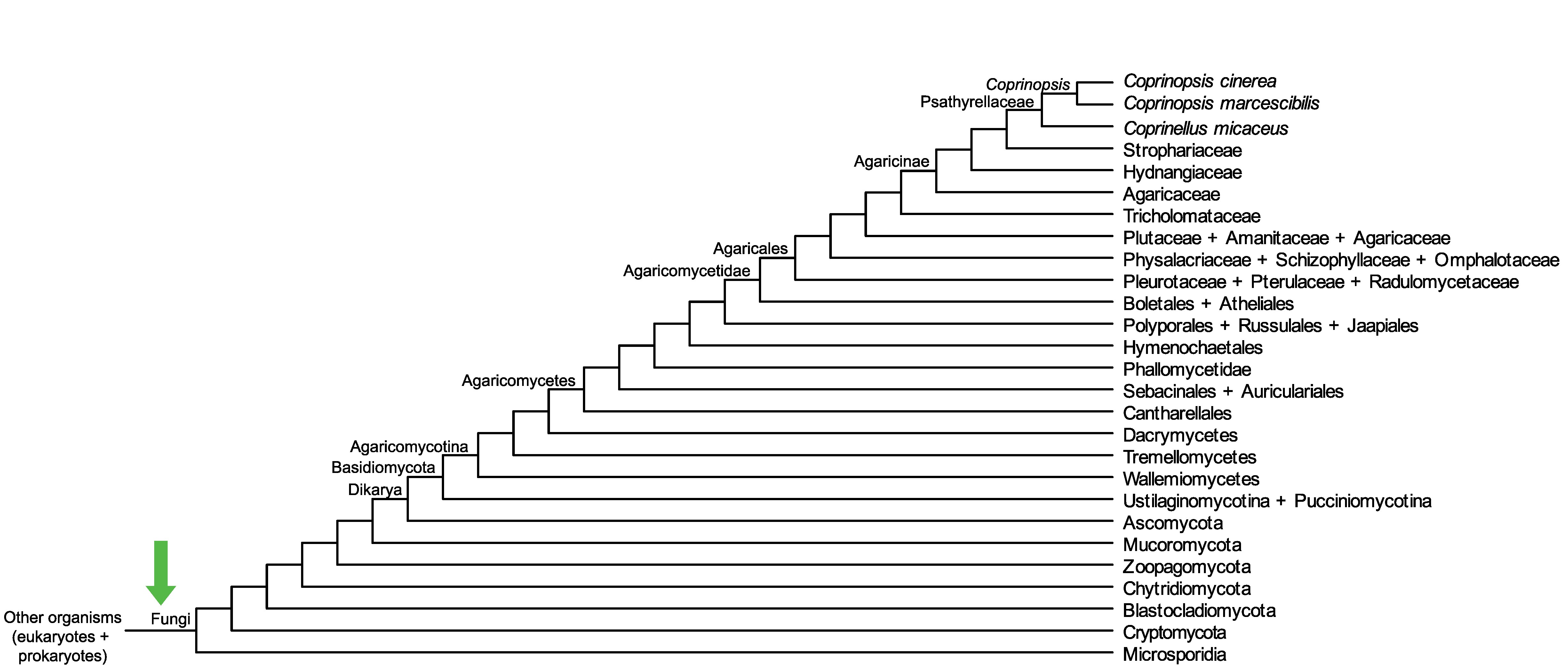
Conservation of CopciAB_443321 across fungi.
Arrow shows the origin of gene family containing CopciAB_443321.
Protein
| Sequence id | CopciAB_443321.T0 |
|---|---|
| Sequence |
>CopciAB_443321.T0 MSWLRGKKSVSNLKASEDLSRATTPTPENPNGKQAKPRFRSGLLTIRVLWAEGLALPAGVAVPTVVSNALSSQQA KVAASVSPSSVHQQRLAKKSKGNRDSVQRTQCWWLPYLVMEYEVNQVLITPLGGDLEKPLYMYQAHFDVSRNSEI SLQCYLRTEEPNCAGDGLADNLGNDIFLGGIKFVPNFDDLGSQDQWYDFVGGSGKLQLGVSYQPNYGQSLTIDDF ELMTVIGKGSFGKVMQVRKRDTSRIYALKTIRKAHIVSRNEITHTLAERLVLAQVDSPFIVPLKFSFQSEQKLYL VLAFVNGGELFHHLQREHTFNEERARFYSAELLLALEHLHELDVVYRDLKPENILLDYTGHIALCDFGLCKLNMK ENEKTNTFCGTPEYLAPEILSGNGYDKSIDWWTLGVLLYEMIHGLPPFYDENTDKMYEKILNKPLEFGDEFSEEA RSILTGLLNRDPSRRLGVKGAEDIKRHPFFHNHIDFKLLAQKKIRPPFKPSVVSPVDVSNFDTVFTEEAPIDSYV ENSNLSSTVQAQFSGFSFNGTNLPSTTP |
| Length | 553 |
Coding
| Sequence id | CopciAB_443321.T0 |
|---|---|
| Sequence |
>CopciAB_443321.T0 ATGTCCTGGCTCCGCGGCAAAAAGTCTGTCAGCAACCTCAAGGCTTCTGAAGACCTTTCACGAGCGACCACACCC ACACCCGAGAACCCCAATGGCAAACAGGCAAAGCCGAGATTTAGGTCGGGTTTGCTGACGATTCGTGTGCTGTGG GCGGAGGGATTGGCTTTGCCAGCTGGCGTGGCCGTCCCGACCGTTGTCAGTAACGCACTGTCATCACAACAAGCC AAGGTTGCTGCCTCTGTCTCGCCCTCCAGCGTACACCAACAGCGACTCGCGAAGAAGTCCAAGGGGAACAGGGAC AGCGTGCAGCGAACACAATGCTGGTGGCTCCCCTACCTCGTCATGGAATACGAAGTCAATCAAGTCCTCATTACA CCTTTGGGTGGTGATCTGGAGAAGCCATTGTACATGTATCAAGCCCATTTCGATGTCTCACGAAACTCGGAAATC TCGTTGCAATGTTACCTCCGAACGGAAGAGCCCAACTGTGCAGGTGACGGCCTCGCAGACAACCTCGGAAACGAT ATCTTCTTGGGTGGCATCAAGTTCGTCCCCAACTTTGACGACTTGGGCAGCCAGGATCAGTGGTATGACTTTGTC GGAGGATCTGGAAAGTTGCAGTTGGGCGTTTCGTACCAACCCAACTATGGTCAATCCCTCACCATCGACGACTTC GAGTTGATGACCGTCATCGGCAAGGGCAGCTTCGGCAAGGTCATGCAAGTTCGCAAACGCGACACGAGCCGGATC TACGCACTGAAGACGATCAGGAAGGCGCACATTGTGTCCCGCAATGAAATTACACATACCCTTGCTGAGCGTCTG GTGTTGGCTCAGGTCGACAGCCCATTCATCGTGCCCCTGAAATTTAGCTTCCAGTCTGAACAGAAGCTCTACCTC GTCCTCGCCTTCGTAAACGGTGGTGAGCTGTTCCATCATCTCCAACGAGAACATACCTTCAACGAGGAGCGGGCG CGGTTCTACAGCGCCGAGCTTCTCCTGGCGCTCGAACATCTCCATGAACTTGATGTTGTTTATCGTGATCTGAAA CCGGAGAACATCCTCCTTGATTATACCGGCCACATCGCCCTCTGTGACTTCGGATTGTGCAAGCTGAACATGAAG GAGAATGAGAAGACGAATACTTTCTGTGGAACACCTGAGTATCTCGCTCCAGAGATTCTGTCGGGCAATGGCTAT GATAAATCCATTGATTGGTGGACTTTGGGTGTCCTGCTCTACGAGATGATCCACGGCCTCCCTCCATTCTACGAT GAAAACACTGACAAGATGTACGAGAAGATCTTGAACAAGCCTCTCGAGTTTGGAGATGAGTTCAGCGAAGAAGCA AGGAGCATCTTGACCGGGCTGCTGAACCGTGATCCCTCTCGCCGCCTCGGTGTTAAGGGAGCGGAGGACATTAAG CGCCATCCCTTCTTCCATAATCATATTGATTTCAAGTTGTTGGCTCAAAAGAAGATTCGTCCTCCGTTCAAGCCC AGCGTTGTGAGCCCTGTCGACGTATCGAACTTTGATACTGTCTTCACGGAAGAAGCGCCGATTGACAGTTATGTG GAGAATTCAAATCTGTCGTCGACTGTCCAGGCTCAGTTCTCTGGCTTCTCGTTCAACGGTACCAACCTTCCCTCT ACTACCCCCTAA |
| Length | 1662 |
Transcript
| Sequence id | CopciAB_443321.T0 |
|---|---|
| Sequence |
>CopciAB_443321.T0 AAACACTTTTGCGCTCAACCACCACCAGCAACAGTCAATTTTGCCTCTGATCCACGACAAATACCCAGCTCAAAC GTCCAGAAAGCAGGCTTTCTCTCATTCCCGCACGCTCCTTAATTCCAATACCGATTAATCACCCACAGAAGCTTA ATCCGTTGCACACCCGACACACACCATGTCCTGGCTCCGCGGCAAAAAGTCTGTCAGCAACCTCAAGGCTTCTGA AGACCTTTCACGAGCGACCACACCCACACCCGAGAACCCCAATGGCAAACAGGCAAAGCCGAGATTTAGGTCGGG TTTGCTGACGATTCGTGTGCTGTGGGCGGAGGGATTGGCTTTGCCAGCTGGCGTGGCCGTCCCGACCGTTGTCAG TAACGCACTGTCATCACAACAAGCCAAGGTTGCTGCCTCTGTCTCGCCCTCCAGCGTACACCAACAGCGACTCGC GAAGAAGTCCAAGGGGAACAGGGACAGCGTGCAGCGAACACAATGCTGGTGGCTCCCCTACCTCGTCATGGAATA CGAAGTCAATCAAGTCCTCATTACACCTTTGGGTGGTGATCTGGAGAAGCCATTGTACATGTATCAAGCCCATTT CGATGTCTCACGAAACTCGGAAATCTCGTTGCAATGTTACCTCCGAACGGAAGAGCCCAACTGTGCAGGTGACGG CCTCGCAGACAACCTCGGAAACGATATCTTCTTGGGTGGCATCAAGTTCGTCCCCAACTTTGACGACTTGGGCAG CCAGGATCAGTGGTATGACTTTGTCGGAGGATCTGGAAAGTTGCAGTTGGGCGTTTCGTACCAACCCAACTATGG TCAATCCCTCACCATCGACGACTTCGAGTTGATGACCGTCATCGGCAAGGGCAGCTTCGGCAAGGTCATGCAAGT TCGCAAACGCGACACGAGCCGGATCTACGCACTGAAGACGATCAGGAAGGCGCACATTGTGTCCCGCAATGAAAT TACACATACCCTTGCTGAGCGTCTGGTGTTGGCTCAGGTCGACAGCCCATTCATCGTGCCCCTGAAATTTAGCTT CCAGTCTGAACAGAAGCTCTACCTCGTCCTCGCCTTCGTAAACGGTGGTGAGCTGTTCCATCATCTCCAACGAGA ACATACCTTCAACGAGGAGCGGGCGCGGTTCTACAGCGCCGAGCTTCTCCTGGCGCTCGAACATCTCCATGAACT TGATGTTGTTTATCGTGATCTGAAACCGGAGAACATCCTCCTTGATTATACCGGCCACATCGCCCTCTGTGACTT CGGATTGTGCAAGCTGAACATGAAGGAGAATGAGAAGACGAATACTTTCTGTGGAACACCTGAGTATCTCGCTCC AGAGATTCTGTCGGGCAATGGCTATGATAAATCCATTGATTGGTGGACTTTGGGTGTCCTGCTCTACGAGATGAT CCACGGCCTCCCTCCATTCTACGATGAAAACACTGACAAGATGTACGAGAAGATCTTGAACAAGCCTCTCGAGTT TGGAGATGAGTTCAGCGAAGAAGCAAGGAGCATCTTGACCGGGCTGCTGAACCGTGATCCCTCTCGCCGCCTCGG TGTTAAGGGAGCGGAGGACATTAAGCGCCATCCCTTCTTCCATAATCATATTGATTTCAAGTTGTTGGCTCAAAA GAAGATTCGTCCTCCGTTCAAGCCCAGCGTTGTGAGCCCTGTCGACGTATCGAACTTTGATACTGTCTTCACGGA AGAAGCGCCGATTGACAGTTATGTGGAGAATTCAAATCTGTCGTCGACTGTCCAGGCTCAGTTCTCTGGCTTCTC GTTCAACGGTACCAACCTTCCCTCTACTACCCCCTAAAGTTGAGCTTTGAGCTCAGCTCGTCATCAACATCATCA CCGTGAACTCACCCTGCAACGTACGCATCTCGCATTCCTGCCTGTTCGAGCTTCCAACAATTCCCTTTTGCTGTG CATTGTACCCCGTCGTATGCTTTCGGTCAGCTTCGCTACGCTCCCTATTATCTTTCTTAATAATCCCACTCCTTT CTCCCCTTAATTTGTATCGCGCTTACTGCTATTATTAATCGCTGATGTTGTGTCCTGTTTTCGAGTACCGTGGCC TTGGTATTACTATTGTGACATCTATACAGTTGTTGAATACAGTACTGGAAAGAAGATCCCTG |
| Length | 2162 |
Gene
| Sequence id | CopciAB_443321.T0 |
|---|---|
| Sequence |
>CopciAB_443321.T0 AAACACTTTTGCGCTCAACCACCACCAGCAACAGTCAATTTTGCCTCTGATCCACGACAAATACCCAGCTCAAAC GTCCAGAAAGCAGGCTTTCTCTCATTCCCGCACGCTCCTTAATTCCAATACCGATTAATCACCCACAGAAGCTTA ATCCGTTGCACACCCGACACACACCATGTCCTGGCTCCGCGGCAAAAAGTCTGTCAGCAACCTCAAGGCTTCTGA AGACCTTTCACGAGCGACCACACCCACACCCGAGAACCCCAATGGCAAACAGGCAAAGCCGAGATTTAGGTCGGG TTTGCTGACGATTCGTGTGCTGTGGGCGGAGGGATTGGCTTTGCCAGCTGGCGTGGCCGTCCCGACCGTTGTCAG TAACGCACTGTCATCACAACAAGCCAAGGTTGCTGCCTCTGTCTCGCCCTCCAGCGTACACCAACAGCGACTCGC GAAGAAGTCCAAGGGGAACAGGTTCGTGTTGCTGATGGACATGTGCTTCGGGATCGTCAGGAGTTAACGACTGCA TTTCAGGGACAGCGTGCAGCGAACACAATGCTGGTGGCTCCCCTACCTCGTCATGGAATACGAAGTCAATCAAGT CCTCATTACACCTTTGGGTGGTGATCTGGAGAAGCCATTGTACATGTATCAAGCCCATTTGTGAGTTCTAATCCT CCAACACACGAGGACGATCACCCCTGATTTAAATTACTTTCCACAGCGATGTCTCACGAAACTCGGAAATCTCGT TGCAATGTTACCTCCGAACGGAAGAGCCCAACTGTGCAGGTGACGGCCTCGCAGACAACCTCGGAAACGATATCT TCTTGGGTGGCATCAAGTTCGTCCCCAACTTTGACGACTTGGGCAGCCAGGATCAGTGGTATGACTTTGTCGGAG GATCTGGAAAGTTGCAGTTGGGCGTTTCGTACCAACCCAACTATGGTCAATCCCTCACCATCGACGACTTCGAGT TGATGACCGTCATCGGCAAGGGCAGCTTCGGCAAGGTCATGCAAGTTCGCAAACGCGACACGAGCCGGATCTACG CACTGAAGACGATCAGGAAGGCGCACATTGTGTCCCGCAATGAAATTACACATACCCTTGCTGAGCGTCTGGTGT TGGCTCAGGTCGACAGCCCATTCATCGTGCCCCTGAAATTTAGCTTCCAGTCTGAACAGAAGCTCTACCTCGTCC TCGCGTAAGTCCTCGTTCTTATGGAATTGGAACGCGTTTTGACCGCATCTTTTTCGCAGCTTCGTAAACGGTGGT GAGCTGTTCCATCATCTCCAACGAGAACATACCTTCAACGAGGAGCGGGCGCGGTACGTGGAGTCGTCGTTGCCA CCACACCTGAAACCTTACGTTCCCTCTAGGTTCTACAGCGCCGAGCTTCTCCTGGCGCTCGAACATCTCCATGAA CTTGATGTTGTTTATCGGTAGGTTTCGTTTCGCCCTTCCATGTGGCCTCGAACGTTAATCTCGTCCCCTGCCGTA GTGATCTGAAACCGGAGAACATCCTCCTTGATTATACCGGCCACATCGCCCTCTGTGACTTCGGATTGTGCAAGC TGAACATGAAGGAGAATGAGAAGACGAATACTTTCTGTGGAACACCTGAGTATCTCGCTCCAGAGATTCTGTCGG GCAATGGCTATGGTGAGACGACCTCTTCCTAGTGTGTCATTGTGCTCGTCCCTGACATTGTCCTTAAGATAAATC CATTGATTGGTGGACTTTGGGTGTCCTGCTCTACGAGATGATCCACGGCCTCCCTCCATTCTACGATGGTAAACC TTTGCACCCTTTGTATACGGACGAGTGCTAACACCGCATATTTGGCGTGCAGAAAACACTGACAAGATGTACGAG AAGATCTTGAACAAGCCTCTCGAGTTTGGAGATGAGTTCAGCGAAGAAGCAAGGAGCATCTTGACCGGGCTGCTG AACCGTGATCCCTCTCGCCGCCTCGGTGTTAAGGGAGCGGAGGACATTAAGCGCCATCCCTTCTTCCATAATCAT ATTGATTTCAAGTTGTTGGCTCAAAAGAAGATTCGTCCTCCGTTCAAGCCCAGCGTTGTGAGCCCTGTCGTAAGT GGTTGTACAATATGCGCCATCGTGTGGATGTGTAACCTGTTGTTTTAGGACGTATCGAACTTTGATACTGTCTTC ACGGAAGAAGCGCCGATTGACAGTTATGTGGAGAATTCAAATCTGTCGTCGACTGTCCAGGCTCAGTTCTCTGGT ATGCCCACCTCTTATCCTCCTTATCGTCGTGAACACGCGGCTGACATATCCTCGCGCGTCAGGCTTCTCGTTCAA CGGTACCAACCTTCCCTCTACTACCCCCTAAAGTTGAGCTTTGAGCTCAGCTCGTCATCAACATCATCACCGTGA ACTCACCCTGCAACGTACGCATCTCGCATTCCTGCCTGTTCGAGCTTCCAACAATTCCCTTTTGCTGTGCATTGT ACCCCGTCGTATGCTTTCGGTCAGCTTCGCTACGCTCCCTATTATCTTTCTTAATAATCCCACTCCTTTCTCCCC TTAATTTGTATCGCGCTTACTGCTATTATTAATCGCTGATGTTGTGTCCTGTTTTCGAGTACCGTGGCCTTGGTA TTACTATTGTGACATCGTGAGTCTACTTCCTGATCTCTTACTAACAGCGGGCGGGCACTGAATGCTTTCACAGTA TACAGTTGTTGAATACAGTACTGGAAAGAAGATCCCTG |
| Length | 2738 |