CopciAB_444045
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_444045 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 444045 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_7:1154198..1157154 | Strand | + |
| Gene length (nt) | 2957 | Transcript length (nt) | 2898 |
| CDS length (nt) | 2382 | Protein length (aa) | 793 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YKL171W_NNK1 |
| Aspergillus nidulans | AN1097_nnk1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7261472 | 50.4 | 2.499E-191 | 614 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14698 | 44 | 6.201E-191 | 613 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_144150 | 46 | 4.055E-152 | 499 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_20125 | 46.5 | 6.053E-150 | 493 |
| Lentinula edodes NBRC 111202 | Lenedo1_1091521 | 46.5 | 6.431E-149 | 490 |
| Grifola frondosa | Grifr_OBZ71123 | 46.7 | 9.254E-140 | 463 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_127520 | 40.2 | 4.438E-139 | 461 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_39794 | 59.6 | 1.023E-133 | 445 |
| Schizophyllum commune H4-8 | Schco3_2459580 | 57.3 | 1.34E-115 | 392 |
| Flammulina velutipes | Flave_chr11AA00042 | 42 | 3.224E-100 | 346 |
| Auricularia subglabra | Aurde3_1_1383316 | 38.8 | 1.271E-92 | 324 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1021681 | 45.8 | 1.443E-90 | 317 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1458726 | 44.6 | 1.448E-79 | 284 |
| Lentinula edodes B17 | Lened_B_1_1_7203 | 55.1 | 8.938E-73 | 263 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87551 | 35.7 | 8.02E-33 | 137 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_444045.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 127 | 281 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 350 | 446 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
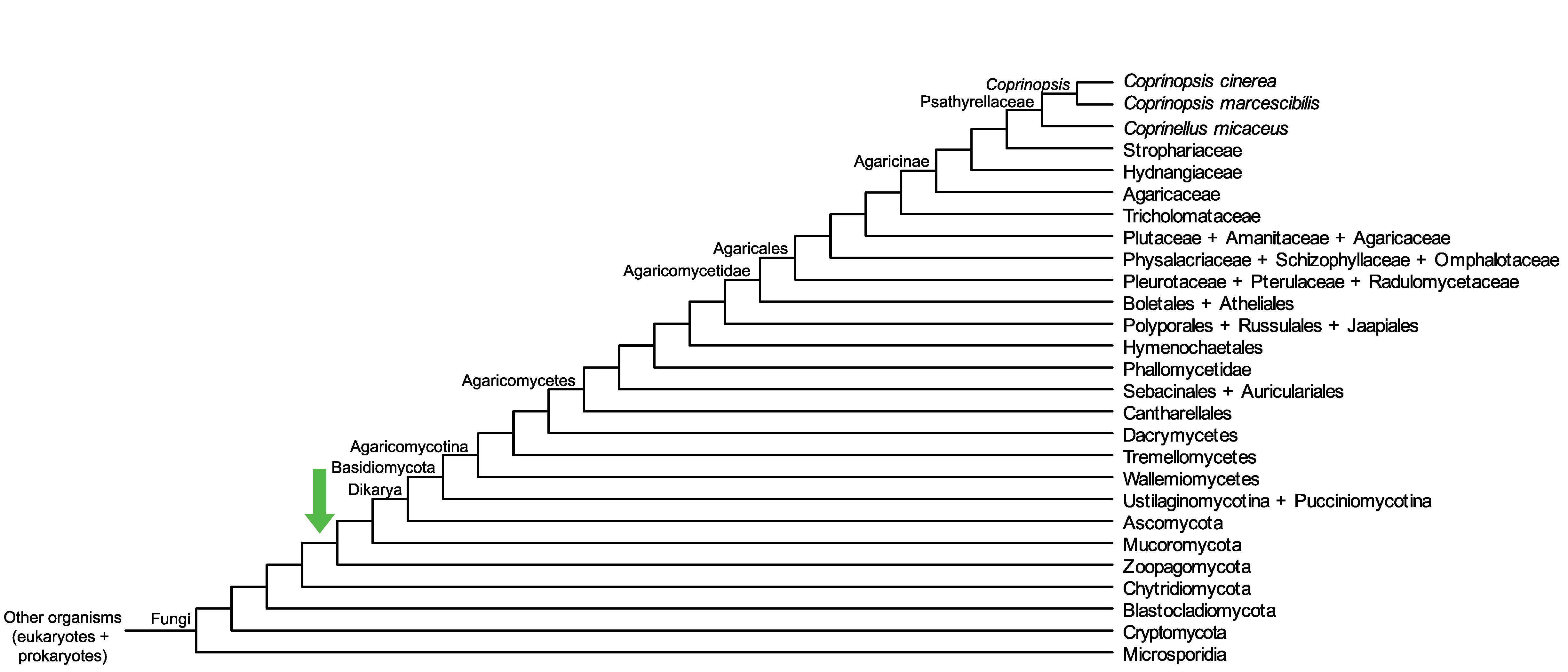
Conservation of CopciAB_444045 across fungi.
Arrow shows the origin of gene family containing CopciAB_444045.
Protein
| Sequence id | CopciAB_444045.T0 |
|---|---|
| Sequence |
>CopciAB_444045.T0 MHSGRIQPQAWSTFANFFPHGDAPHLATSPLPVNESHGHFPSPQKVGDEDVLGDSELDKEQRVSAAFIDADTIIE DHSPSISPVRPSVSQPTTLASSPAALFLSAFMGSKPEPEPLEDDEGQKVGSYTLGKVVGFGAFSTIRTGVSESGS TVAVKIVRRADLVRTGHAPEERKRLQHEASIWSSLSHEHILPLFSALHTSYADYFVTIYCPAGSLYDILKRDGRP ALPQDDAGMMFRQVVRGLRYLHEVAGLVHRDIKLENVLVDESGVCRIGDFGLSVRIGDVDDEDGMDHHHHEHHPI DGACTGNIHRAVSLSFPSKRHRAATTLGAAIARHNSTRNRHAGQSSHVYQPGSLPYAAPELLLPNTADMAPAHPG QDIWALGVMLYALLTGHLPFNDSFEPRLQMKILNGTFEIPADIGRGAERILQGCMERIITKRWTIAMVDEVSWGV GWGSAGDNATPDESQEDLLAFHAITSPLSAKSCSESRLDRLTIPESDEDDWQHEEHHFTRSAIEAGSRRSSSRAK RSQSRAPMSARSHSRQCSPHRSARSSSRMYSGFSSAAVSRSHSPATSPLFENPPSRSRSQSRHRGRQPNKRCYFA SRSPSPSWVPSTPSDISHISRLSLREPSEDLHLDSPPRGRTRFSLHHETFDEGDEATEGVEGGGALKLTHAADGT GQCSATDWTSRITAEAEDHVSPERASSRPNIRSSWTEIPFRNSRPGSTPPASSKTWELFRNKASHGLLSRNPSQD MCLGFPSPGLGTPGIISRSRSIDQYHDPTRTAKVEIPPASSCA |
| Length | 793 |
Coding
| Sequence id | CopciAB_444045.T0 |
|---|---|
| Sequence |
>CopciAB_444045.T0 ATGCATAGTGGTCGAATCCAGCCTCAAGCTTGGTCCACCTTCGCTAATTTCTTTCCTCACGGGGATGCTCCACAC CTTGCCACTTCTCCGCTTCCAGTAAACGAGAGTCATGGCCACTTCCCAAGCCCGCAAAAAGTCGGCGATGAGGAC GTATTGGGGGATTCAGAGCTTGACAAGGAGCAGAGGGTCTCTGCGGCCTTCATAGACGCGGACACCATAATCGAG GACCATTCACCATCAATCTCACCTGTTCGCCCGAGCGTCTCACAACCAACAACCCTCGCCAGTTCCCCAGCAGCT CTCTTTCTGTCAGCCTTCATGGGCTCCAAGCCTGAGCCCGAGCCTCTCGAAGACGATGAAGGCCAGAAAGTCGGG TCTTACACCCTCGGAAAGGTAGTTGGATTTGGCGCCTTTTCTACCATTAGAACGGGTGTCTCTGAGTCGGGCTCC ACCGTGGCAGTCAAGATCGTCCGCAGAGCTGACCTCGTTCGAACCGGTCACGCCCCAGAAGAACGCAAGAGACTG CAACACGAAGCCTCGATATGGTCTTCACTCAGCCATGAACATATCCTACCTTTATTTTCAGCACTCCACACTTCC TATGCCGACTACTTCGTCACAATCTACTGCCCAGCCGGCTCTCTGTACGACATCCTCAAAAGGGACGGACGTCCT GCGTTGCCCCAAGACGACGCAGGGATGATGTTTCGGCAGGTGGTGCGGGGGTTGCGGTATTTGCATGAAGTTGCG GGGTTGGTCCATAGGGACATCAAACTAGAAAATGTTTTAGTGGATGAATCAGGTGTTTGTAGAATTGGTGACTTT GGATTGAGTGTCAGGATTGGTGACGTGGACGACGAGGATGGCATGGACCACCATCACCATGAACACCATCCCATT GATGGAGCTTGCACCGGTAACATTCATCGTGCTGTATCTCTGTCCTTCCCATCCAAGCGCCACAGAGCTGCGACC ACCCTCGGTGCAGCTATCGCTCGCCACAATAGCACTCGCAATCGACATGCTGGCCAAAGCTCACATGTCTATCAG CCCGGTTCCCTTCCCTACGCCGCGCCCGAACTTCTCCTGCCCAACACAGCGGATATGGCCCCCGCCCATCCTGGC CAGGATATATGGGCGCTTGGAGTTATGCTTTATGCTTTGTTGACTGGTCATTTGCCGTTCAATGATTCGTTTGAG CCTCGCTTGCAGATGAAAATCCTAAATGGTACTTTTGAGATACCAGCCGACATCGGTCGCGGTGCTGAGCGCATT CTCCAAGGATGCATGGAGCGCATAATCACCAAGCGATGGACCATCGCCATGGTCGATGAAGTGTCTTGGGGCGTT GGATGGGGATCAGCCGGCGACAACGCCACACCAGACGAATCGCAAGAGGACCTACTTGCATTCCATGCCATCACA TCCCCCCTCTCCGCCAAGTCATGTTCAGAGTCTCGCCTCGACAGACTCACCATCCCCGAATCAGATGAAGACGAC TGGCAACACGAAGAGCACCATTTCACTCGGTCCGCAATTGAGGCGGGCTCCAGGCGATCGTCCTCGCGGGCGAAA CGCTCGCAATCCCGCGCACCAATGTCAGCTCGCAGCCACTCTCGACAGTGCTCTCCTCACCGTAGTGCACGGAGC AGCTCTCGAATGTATTCTGGGTTTTCATCTGCTGCCGTGTCTCGGAGCCATTCGCCTGCAACATCGCCGTTATTC GAAAATCCTCCCAGCAGGAGCCGCTCACAGTCCCGGCACCGTGGGCGACAGCCCAACAAGCGGTGTTATTTCGCC TCTCGCTCCCCTAGCCCCAGTTGGGTCCCTTCAACGCCCAGCGACATCAGCCACATTTCAAGGTTGAGCCTACGC GAACCATCGGAGGACCTGCATCTTGACTCACCTCCCCGCGGTCGAACTCGGTTCTCTTTGCACCACGAAACATTC GACGAAGGAGACGAAGCGACAGAAGGAGTGGAGGGAGGGGGAGCATTGAAGCTCACACACGCAGCCGACGGCACA GGTCAATGTTCGGCCACCGACTGGACGTCTCGTATCACTGCTGAGGCAGAGGACCACGTCTCCCCAGAAAGAGCC AGCTCCCGTCCGAATATCCGCTCGTCGTGGACCGAGATCCCCTTCCGCAACTCGAGACCGGGTAGCACCCCTCCT GCGTCGTCCAAGACTTGGGAACTGTTCAGGAACAAGGCGTCGCATGGCCTGTTGTCGCGGAATCCCTCCCAGGAC ATGTGCCTGGGGTTTCCGTCGCCTGGCCTGGGAACTCCTGGCATCATATCTCGAAGTCGTAGTATCGATCAATAT CATGACCCGACCCGCACTGCAAAAGTTGAGATACCTCCCGCATCAAGTTGCGCATGA |
| Length | 2382 |
Transcript
| Sequence id | CopciAB_444045.T0 |
|---|---|
| Sequence |
>CopciAB_444045.T0 AGTTCCTCTTTCCCTTTAACCATCACCACAGCCAACTGTCGTCCACACAATTTATCTTCTCTGCTCCCCAGATTC ACATAGTATCGTCGACAAGCCCGTTACATGCATAGTGGTCGAATCCAGCCTCAAGCTTGGTCCACCTTCGCTAAT TTCTTTCCTCACGGGGATGCTCCACACCTTGCCACTTCTCCGCTTCCAGTAAACGAGAGTCATGGCCACTTCCCA AGCCCGCAAAAAGTCGGCGATGAGGACGTATTGGGGGATTCAGAGCTTGACAAGGAGCAGAGGGTCTCTGCGGCC TTCATAGACGCGGACACCATAATCGAGGACCATTCACCATCAATCTCACCTGTTCGCCCGAGCGTCTCACAACCA ACAACCCTCGCCAGTTCCCCAGCAGCTCTCTTTCTGTCAGCCTTCATGGGCTCCAAGCCTGAGCCCGAGCCTCTC GAAGACGATGAAGGCCAGAAAGTCGGGTCTTACACCCTCGGAAAGGTAGTTGGATTTGGCGCCTTTTCTACCATT AGAACGGGTGTCTCTGAGTCGGGCTCCACCGTGGCAGTCAAGATCGTCCGCAGAGCTGACCTCGTTCGAACCGGT CACGCCCCAGAAGAACGCAAGAGACTGCAACACGAAGCCTCGATATGGTCTTCACTCAGCCATGAACATATCCTA CCTTTATTTTCAGCACTCCACACTTCCTATGCCGACTACTTCGTCACAATCTACTGCCCAGCCGGCTCTCTGTAC GACATCCTCAAAAGGGACGGACGTCCTGCGTTGCCCCAAGACGACGCAGGGATGATGTTTCGGCAGGTGGTGCGG GGGTTGCGGTATTTGCATGAAGTTGCGGGGTTGGTCCATAGGGACATCAAACTAGAAAATGTTTTAGTGGATGAA TCAGGTGTTTGTAGAATTGGTGACTTTGGATTGAGTGTCAGGATTGGTGACGTGGACGACGAGGATGGCATGGAC CACCATCACCATGAACACCATCCCATTGATGGAGCTTGCACCGGTAACATTCATCGTGCTGTATCTCTGTCCTTC CCATCCAAGCGCCACAGAGCTGCGACCACCCTCGGTGCAGCTATCGCTCGCCACAATAGCACTCGCAATCGACAT GCTGGCCAAAGCTCACATGTCTATCAGCCCGGTTCCCTTCCCTACGCCGCGCCCGAACTTCTCCTGCCCAACACA GCGGATATGGCCCCCGCCCATCCTGGCCAGGATATATGGGCGCTTGGAGTTATGCTTTATGCTTTGTTGACTGGT CATTTGCCGTTCAATGATTCGTTTGAGCCTCGCTTGCAGATGAAAATCCTAAATGGTACTTTTGAGATACCAGCC GACATCGGTCGCGGTGCTGAGCGCATTCTCCAAGGATGCATGGAGCGCATAATCACCAAGCGATGGACCATCGCC ATGGTCGATGAAGTGTCTTGGGGCGTTGGATGGGGATCAGCCGGCGACAACGCCACACCAGACGAATCGCAAGAG GACCTACTTGCATTCCATGCCATCACATCCCCCCTCTCCGCCAAGTCATGTTCAGAGTCTCGCCTCGACAGACTC ACCATCCCCGAATCAGATGAAGACGACTGGCAACACGAAGAGCACCATTTCACTCGGTCCGCAATTGAGGCGGGC TCCAGGCGATCGTCCTCGCGGGCGAAACGCTCGCAATCCCGCGCACCAATGTCAGCTCGCAGCCACTCTCGACAG TGCTCTCCTCACCGTAGTGCACGGAGCAGCTCTCGAATGTATTCTGGGTTTTCATCTGCTGCCGTGTCTCGGAGC CATTCGCCTGCAACATCGCCGTTATTCGAAAATCCTCCCAGCAGGAGCCGCTCACAGTCCCGGCACCGTGGGCGA CAGCCCAACAAGCGGTGTTATTTCGCCTCTCGCTCCCCTAGCCCCAGTTGGGTCCCTTCAACGCCCAGCGACATC AGCCACATTTCAAGGTTGAGCCTACGCGAACCATCGGAGGACCTGCATCTTGACTCACCTCCCCGCGGTCGAACT CGGTTCTCTTTGCACCACGAAACATTCGACGAAGGAGACGAAGCGACAGAAGGAGTGGAGGGAGGGGGAGCATTG AAGCTCACACACGCAGCCGACGGCACAGGTCAATGTTCGGCCACCGACTGGACGTCTCGTATCACTGCTGAGGCA GAGGACCACGTCTCCCCAGAAAGAGCCAGCTCCCGTCCGAATATCCGCTCGTCGTGGACCGAGATCCCCTTCCGC AACTCGAGACCGGGTAGCACCCCTCCTGCGTCGTCCAAGACTTGGGAACTGTTCAGGAACAAGGCGTCGCATGGC CTGTTGTCGCGGAATCCCTCCCAGGACATGTGCCTGGGGTTTCCGTCGCCTGGCCTGGGAACTCCTGGCATCATA TCTCGAAGTCGTAGTATCGATCAATATCATGACCCGACCCGCACTGCAAAAGTTGAGATACCTCCCGCATCAAGT TGCGCATGATGTGCTTACCGATGGATCTTTCTTGCTCATCTAACATCCCCCGTCCCTATTTATGCGTCTTATGGT TTTCCGACGGTAGTTGATTCTTTTTGCTTCTGAGACGTTTCTTCAGTGGTCAAGCAGCGTCGTTGTAGCATTAGC ACTTTACTAACCTAGGTTCCGCTTTTAAAACCCTTTTCCTCCTTTCCGCATCTTCTCATTTCTTCTGGTTACTAG GCTTTCTTCTTGAGTCATCTGAAGTTGGTCTTTTCCTGGTCTGCCGTTCATTCTGTACTCTATGTTGGTTGGTTC CGAGTTCGTGCATTGGTACTTTTCCAAAAACAATCGGGATACCCTCATGTTGTAGCAATACGTCTTTCCCTTCAT TCCTGTTCAGTATTCACTTGTGGTCCCAAATGCCATCCGTTTTTCACG |
| Length | 2898 |
Gene
| Sequence id | CopciAB_444045.T0 |
|---|---|
| Sequence |
>CopciAB_444045.T0 AGTTCCTCTTTCCCTTTAACCATCACCACAGCCAACTGTCGTCCACACAATTTATCTTCTCTGCTCCCCAGATTC ACATAGTATCGTCGACAAGCCCGTTACATGCATAGTGGTCGAATCCAGCCTCAAGCTTGGTCCACCTTCGCTAAT TTCTTTCCTCACGGGGATGCTCCACACCTTGCCACTTCTCCGCTTCCAGTAAACGAGAGTCATGGCCACTTCCCA AGCCCGCAAAAAGTCGGCGATGAGGACGTATTGGGGGATTCAGAGCTTGACAAGGAGCAGAGGGTCTCTGCGGCC TTCATAGACGCGGACACCATAATCGAGGACCATTCACCATCAATCTCACCTGTTCGCCCGAGCGTCTCACAACCA ACAACCCTCGCCAGTTCCCCAGCAGCTCTCTTTCTGTCAGCCTTCATGGGCTCCAAGCCTGAGCCCGAGCCTCTC GAAGACGATGAAGGCCAGAAAGTCGGGTCTTACACCCTCGGAAAGGTAGTTGGATTTGGCGCCTTTTCTACCATT AGAACGGGTGTCTCTGAGTCGGGCTCCACCGTGGCAGTCAAGATCGTCCGCAGAGCTGACCTCGTTCGAACCGGT CACGCCCCAGAAGAACGCAAGAGACTGCAACACGAAGCCTCGATATGGTCTTCACTCAGCCATGAACATATCCTA CCTTTATTTTCAGCACTCCACACTTCCTATGCCGACTACTTCGTCACAATCTACTGCCCAGCCGGCTCTCTGTAC GACATCCTCAAAAGGGACGGACGTCCTGCGTTGCCCCAAGACGACGCAGGGATGATGTTTCGGCAGGTGGTGCGG GGGTTGCGGTATTTGCATGAAGTTGCGGGGTTGGTCCATAGGGACATCAAACTAGAAAATGTTTTAGTGGATGAA TCAGGTGTTTGTAGAATTGGTGACTTTGGATTGAGTGTCAGGATTGGTGACGTGGACGACGAGGATGGCATGGAC CACCATCACCATGAACACCATCCCATTGATGGAGCTTGCACCGGTAACATTCATCGTGCTGTATCTCTGTCCTTC CCATCCAAGCGCCACAGAGCTGCGACCACCCTCGGTGCAGCTATCGCTCGCCACAATAGCACTCGCAATCGACAT GCTGGCCAAAGCTCACATGTCTATCAGCCCGGTTCCCTTCCCTACGCCGCGCCCGAACTTCTCCTGCCCAACACA GCGGATATGGCCCCCGCCCATCCTGGCCAGGATATATGGGCGCTTGGAGTTATGCTTTATGCTTTGTTGACTGGT CATTTGCCGTTCAATGATTCGTTTGAGCCTCGCTTGCAGATGAAAATCCTAAATGGTACGTATGAACGCTTGATC ACTCGCCTTATGTTGTTTACCCACCGGTGATATCCCTAGGTACTTTTGAGATACCAGCCGACATCGGTCGCGGTG CTGAGCGCATTCTCCAAGGATGCATGGAGCGCATAATCACCAAGCGATGGACCATCGCCATGGTCGATGAAGTGT CTTGGGGCGTTGGATGGGGATCAGCCGGCGACAACGCCACACCAGACGAATCGCAAGAGGACCTACTTGCATTCC ATGCCATCACATCCCCCCTCTCCGCCAAGTCATGTTCAGAGTCTCGCCTCGACAGACTCACCATCCCCGAATCAG ATGAAGACGACTGGCAACACGAAGAGCACCATTTCACTCGGTCCGCAATTGAGGCGGGCTCCAGGCGATCGTCCT CGCGGGCGAAACGCTCGCAATCCCGCGCACCAATGTCAGCTCGCAGCCACTCTCGACAGTGCTCTCCTCACCGTA GTGCACGGAGCAGCTCTCGAATGTATTCTGGGTTTTCATCTGCTGCCGTGTCTCGGAGCCATTCGCCTGCAACAT CGCCGTTATTCGAAAATCCTCCCAGCAGGAGCCGCTCACAGTCCCGGCACCGTGGGCGACAGCCCAACAAGCGGT GTTATTTCGCCTCTCGCTCCCCTAGCCCCAGTTGGGTCCCTTCAACGCCCAGCGACATCAGCCACATTTCAAGGT TGAGCCTACGCGAACCATCGGAGGACCTGCATCTTGACTCACCTCCCCGCGGTCGAACTCGGTTCTCTTTGCACC ACGAAACATTCGACGAAGGAGACGAAGCGACAGAAGGAGTGGAGGGAGGGGGAGCATTGAAGCTCACACACGCAG CCGACGGCACAGGTCAATGTTCGGCCACCGACTGGACGTCTCGTATCACTGCTGAGGCAGAGGACCACGTCTCCC CAGAAAGAGCCAGCTCCCGTCCGAATATCCGCTCGTCGTGGACCGAGATCCCCTTCCGCAACTCGAGACCGGGTA GCACCCCTCCTGCGTCGTCCAAGACTTGGGAACTGTTCAGGAACAAGGCGTCGCATGGCCTGTTGTCGCGGAATC CCTCCCAGGACATGTGCCTGGGGTTTCCGTCGCCTGGCCTGGGAACTCCTGGCATCATATCTCGAAGTCGTAGTA TCGATCAATATCATGACCCGACCCGCACTGCAAAAGTTGAGATACCTCCCGCATCAAGTTGCGCATGATGTGCTT ACCGATGGATCTTTCTTGCTCATCTAACATCCCCCGTCCCTATTTATGCGTCTTATGGTTTTCCGACGGTAGTTG ATTCTTTTTGCTTCTGAGACGTTTCTTCAGTGGTCAAGCAGCGTCGTTGTAGCATTAGCACTTTACTAACCTAGG TTCCGCTTTTAAAACCCTTTTCCTCCTTTCCGCATCTTCTCATTTCTTCTGGTTACTAGGCTTTCTTCTTGAGTC ATCTGAAGTTGGTCTTTTCCTGGTCTGCCGTTCATTCTGTACTCTATGTTGGTTGGTTCCGAGTTCGTGCATTGG TACTTTTCCAAAAACAATCGGGATACCCTCATGTTGTAGCAATACGTCTTTCCCTTCATTCCTGTTCAGTATTCA CTTGTGGTCCCAAATGCCATCCGTTTTTCACG |
| Length | 2957 |