CopciAB_445594
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_445594 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | CDC7 | Synonyms | 445594 |
| Uniprot id | Functional description | Protein kinase domain | |
| Location | scaffold_5:547488..550201 | Strand | - |
| Gene length (nt) | 2714 | Transcript length (nt) | 2437 |
| CDS length (nt) | 2307 | Protein length (aa) | 768 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YDL017W_CDC7 |
| Neurospora crassa | cdc7 |
| Aspergillus nidulans | AN3450_cdc7 |
| Schizosaccharomyces pombe | hsk1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB20032 | 50.3 | 1.161E-223 | 707 |
| Grifola frondosa | Grifr_OBZ70482 | 48.2 | 2.771E-208 | 662 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1055539 | 47.5 | 5.717E-208 | 661 |
| Pleurotus ostreatus PC9 | PleosPC9_1_83213 | 47.5 | 5.595E-208 | 661 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1442992 | 47.4 | 3.525E-206 | 656 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_188660 | 47 | 1.356E-204 | 651 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_72476 | 46.9 | 3.119E-204 | 650 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_82464 | 47.7 | 8.808E-203 | 646 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_15093 | 42.8 | 3.227E-196 | 627 |
| Schizophyllum commune H4-8 | Schco3_2612626 | 44.6 | 7.399E-186 | 597 |
| Agrocybe aegerita | Agrae_CAA7259341 | 42 | 7.293E-185 | 594 |
| Lentinula edodes NBRC 111202 | Lenedo1_1063059 | 48.3 | 6.17E-175 | 565 |
| Flammulina velutipes | Flave_chr11AA01114 | 38.3 | 6.018E-148 | 486 |
| Auricularia subglabra | Aurde3_1_188361 | 37.5 | 1.778E-137 | 456 |
| Lentinula edodes B17 | Lened_B_1_1_13389 | 52.7 | 7.165E-42 | 166 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | CDC7 |
|---|---|
| Protein id | CopciAB_445594.T0 |
| Description | Protein kinase domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14019 | STKc_Cdc7 | - | 198 | 498 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 252 | 374 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 434 | 669 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K02214 |
EggNOG
| COG category | Description |
|---|---|
| L | Protein kinase domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
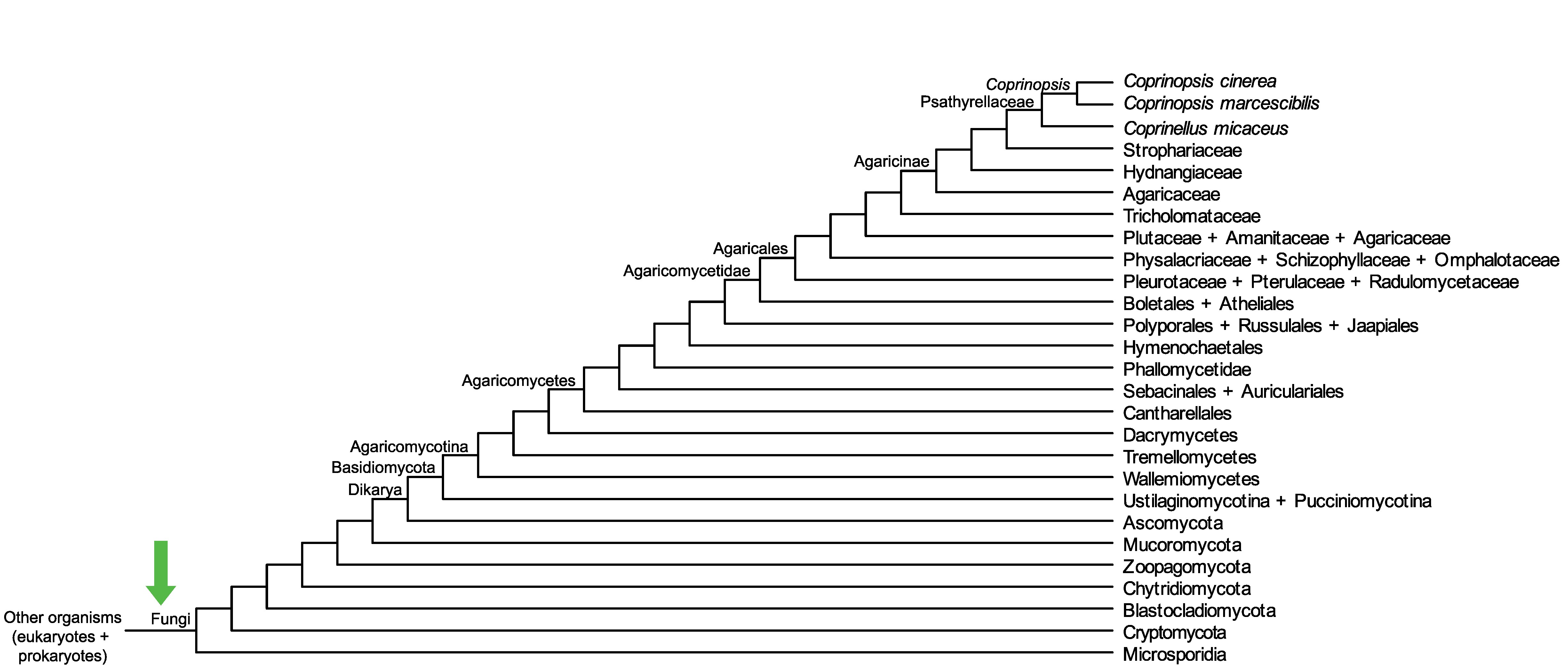
Conservation of CopciAB_445594 across fungi.
Arrow shows the origin of gene family containing CopciAB_445594.
Protein
| Sequence id | CopciAB_445594.T0 |
|---|---|
| Sequence |
>CopciAB_445594.T0 MSVTYFSQHSSNPLEARTSDARNLHYNRVYAEANAQLDEVSPSSDPLARYPQDELREYHEHLAEVIPGHPQTMSQ ARKKETRMVELDEDGDGEEEMSDDELMIIPKEEEDVSVDDGEQDKEGNTPQQQQFAEYDEDMDAEGEWDEDVEVE EPKEKDYFALDESEEMGSIDKKTEDEIAEIEKEIEEFEVALPTLASGYKLLDRLGTGTFSSVYKALDLRYHEYDN SPWLGHHPEESSAHFQSVPQPEGSRVFVAIKRIYVTSGPERIRNELFIMETCRGARHVSQLITAFRHHDQVAIVM PYHRNEDFRDYYETLPMEGIKSYFRCLLRALRDIHARGIIHRDVKPANFLFDPSTGQGTLCDFGLAQIMDDKPPV LGACLHTQPTEQYPHGHIRKKHQYDENLVKEGQMDARKKSKMHAHSVGILDKDTRMPMKANRAGTRGFRAPEVLL KCGQQSGAVDVWSAGIILLFFLTRKFPIFQSNDDIEALMELAVIFGRKKMESVATLHARKFATNVPDIPQDQVKW TTMIEKLNPDLLTPPTPKEHFYPYTLPSYRAKLRQEEERRQQRQQDTHRTPTRSSRHEQYQEDDVESDAAPPTSR LTSSSPLLFPRSSPANHVDEDIPSSPGSDSLQASWEQGIRDAMDLVTKLLEPESVKRITPAAALAHPFLREEDKL DDDYYVPHVFGRGVCGKYHFYDDVTEQPGVIIRERRRKCVCNCGLEECQEQDENAGEWVKVRKLVSAGEGIAIGM EPCEFHQDYQLYDDDEDD |
| Length | 768 |
Coding
| Sequence id | CopciAB_445594.T0 |
|---|---|
| Sequence |
>CopciAB_445594.T0 ATGTCTGTCACTTATTTTTCCCAACACTCTTCTAATCCGCTCGAGGCAAGGACTTCCGATGCCCGCAATCTTCAT TACAACCGCGTCTACGCAGAGGCCAACGCCCAACTCGACGAAGTCTCTCCCTCGAGCGATCCGCTAGCCCGATAT CCTCAAGACGAACTGCGCGAATACCACGAACATCTTGCAGAGGTCATTCCAGGGCACCCTCAGACAATGAGCCAA GCGCGGAAGAAAGAGACACGGATGGTGGAGCTCGACGAGGACGGTGACGGCGAAGAGGAGATGAGTGACGACGAG CTCATGATCATCCCCAAGGAGGAAGAGGACGTCTCCGTCGACGATGGAGAACAGGACAAGGAAGGCAACACCCCA CAGCAACAGCAATTCGCCGAGTACGACGAGGACATGGATGCAGAGGGCGAATGGGACGAGGACGTAGAGGTTGAA GAGCCCAAGGAGAAGGATTATTTCGCCCTGGACGAATCTGAAGAAATGGGCTCGATAGACAAGAAAACCGAAGAC GAAATTGCCGAAATCGAGAAGGAAATCGAAGAGTTTGAAGTGGCCCTTCCGACACTCGCTTCTGGGTACAAACTC CTCGACCGGTTGGGCACTGGGACGTTTTCTTCGGTCTACAAAGCACTCGACCTGCGGTATCACGAATACGACAAC AGTCCATGGCTAGGACACCATCCGGAGGAGTCTTCAGCTCACTTCCAGTCCGTTCCGCAACCAGAGGGGTCGAGG GTATTCGTGGCTATCAAGAGGATTTACGTTACAAGTGGCCCGGAGAGGATACGGAACGAACTGTTCATCATGGAG ACGTGCAGGGGAGCAAGACACGTTTCGCAGCTCATCACCGCGTTCAGGCACCATGACCAGGTCGCCATCGTTATG CCTTACCATCGGAATGAGGATTTCAGGGACTACTACGAGACGCTACCGATGGAAGGAATCAAATCCTATTTCCGA TGTCTACTACGCGCATTACGCGATATCCACGCGAGGGGGATAATCCATCGCGACGTCAAACCAGCCAACTTTTTG TTTGATCCCAGCACCGGTCAGGGGACCCTGTGCGACTTTGGGTTGGCTCAGATCATGGACGACAAGCCTCCTGTG CTAGGCGCATGCCTGCACACGCAGCCCACGGAGCAATACCCCCACGGTCACATCCGGAAGAAACACCAGTACGAC GAGAATCTGGTGAAGGAGGGACAGATGGACGCGCGGAAGAAGAGTAAAATGCATGCGCATTCTGTTGGTATTCTT GACAAAGATACTCGAATGCCCATGAAAGCCAATCGTGCTGGTACTCGAGGATTCCGCGCACCTGAAGTCTTGCTC AAGTGCGGTCAGCAATCCGGAGCTGTGGATGTCTGGTCTGCGGGAATAATCCTCCTCTTCTTCCTCACTCGAAAG TTCCCAATATTCCAATCCAACGACGATATCGAGGCTCTCATGGAACTGGCTGTCATCTTCGGGCGGAAGAAAATG GAGAGCGTAGCCACACTACACGCCCGCAAATTTGCAACCAACGTCCCCGACATTCCACAAGATCAAGTCAAATGG ACGACAATGATCGAGAAGCTCAATCCGGACCTTCTGACACCACCCACTCCGAAAGAGCATTTCTATCCCTACACG CTCCCTTCGTATCGTGCAAAGCTACGACAGGAAGAGGAGCGAAGGCAACAACGTCAACAGGATACTCACAGGACG CCGACAAGGTCCTCAAGGCACGAACAATATCAAGAAGATGACGTCGAGTCCGACGCCGCGCCTCCAACATCCCGC TTAACCTCCTCTTCCCCACTCTTATTCCCTCGATCCTCACCCGCGAACCATGTCGACGAGGACATTCCTTCATCA CCAGGTTCCGACTCTTTACAAGCCTCATGGGAACAAGGCATCCGAGACGCCATGGACCTCGTTACCAAACTCCTC GAACCCGAGTCCGTAAAGAGGATAACACCAGCCGCAGCCCTCGCCCACCCTTTCCTCCGCGAAGAAGATAAACTT GACGATGACTACTATGTTCCACACGTTTTCGGACGAGGTGTTTGTGGCAAGTACCATTTCTATGATGATGTGACG GAGCAGCCGGGGGTTATCATTAGGGAGAGGAGACGGAAGTGTGTCTGTAATTGCGGACTGGAAGAGTGCCAGGAG CAGGATGAGAATGCAGGGGAGTGGGTGAAGGTGAGGAAGCTGGTCTCCGCTGGCGAGGGGATAGCGATTGGGATG GAGCCATGCGAGTTCCACCAAGATTACCAACTTTACGACGATGATGAGGACGATTGA |
| Length | 2307 |
Transcript
| Sequence id | CopciAB_445594.T0 |
|---|---|
| Sequence |
>CopciAB_445594.T0 GGAAATTGGATCTCGTTCTGTCAACGGGCTTCTTTCTTAGCTCAATGTCTGTCACTTATTTTTCCCAACACTCTT CTAATCCGCTCGAGGCAAGGACTTCCGATGCCCGCAATCTTCATTACAACCGCGTCTACGCAGAGGCCAACGCCC AACTCGACGAAGTCTCTCCCTCGAGCGATCCGCTAGCCCGATATCCTCAAGACGAACTGCGCGAATACCACGAAC ATCTTGCAGAGGTCATTCCAGGGCACCCTCAGACAATGAGCCAAGCGCGGAAGAAAGAGACACGGATGGTGGAGC TCGACGAGGACGGTGACGGCGAAGAGGAGATGAGTGACGACGAGCTCATGATCATCCCCAAGGAGGAAGAGGACG TCTCCGTCGACGATGGAGAACAGGACAAGGAAGGCAACACCCCACAGCAACAGCAATTCGCCGAGTACGACGAGG ACATGGATGCAGAGGGCGAATGGGACGAGGACGTAGAGGTTGAAGAGCCCAAGGAGAAGGATTATTTCGCCCTGG ACGAATCTGAAGAAATGGGCTCGATAGACAAGAAAACCGAAGACGAAATTGCCGAAATCGAGAAGGAAATCGAAG AGTTTGAAGTGGCCCTTCCGACACTCGCTTCTGGGTACAAACTCCTCGACCGGTTGGGCACTGGGACGTTTTCTT CGGTCTACAAAGCACTCGACCTGCGGTATCACGAATACGACAACAGTCCATGGCTAGGACACCATCCGGAGGAGT CTTCAGCTCACTTCCAGTCCGTTCCGCAACCAGAGGGGTCGAGGGTATTCGTGGCTATCAAGAGGATTTACGTTA CAAGTGGCCCGGAGAGGATACGGAACGAACTGTTCATCATGGAGACGTGCAGGGGAGCAAGACACGTTTCGCAGC TCATCACCGCGTTCAGGCACCATGACCAGGTCGCCATCGTTATGCCTTACCATCGGAATGAGGATTTCAGGGACT ACTACGAGACGCTACCGATGGAAGGAATCAAATCCTATTTCCGATGTCTACTACGCGCATTACGCGATATCCACG CGAGGGGGATAATCCATCGCGACGTCAAACCAGCCAACTTTTTGTTTGATCCCAGCACCGGTCAGGGGACCCTGT GCGACTTTGGGTTGGCTCAGATCATGGACGACAAGCCTCCTGTGCTAGGCGCATGCCTGCACACGCAGCCCACGG AGCAATACCCCCACGGTCACATCCGGAAGAAACACCAGTACGACGAGAATCTGGTGAAGGAGGGACAGATGGACG CGCGGAAGAAGAGTAAAATGCATGCGCATTCTGTTGGTATTCTTGACAAAGATACTCGAATGCCCATGAAAGCCA ATCGTGCTGGTACTCGAGGATTCCGCGCACCTGAAGTCTTGCTCAAGTGCGGTCAGCAATCCGGAGCTGTGGATG TCTGGTCTGCGGGAATAATCCTCCTCTTCTTCCTCACTCGAAAGTTCCCAATATTCCAATCCAACGACGATATCG AGGCTCTCATGGAACTGGCTGTCATCTTCGGGCGGAAGAAAATGGAGAGCGTAGCCACACTACACGCCCGCAAAT TTGCAACCAACGTCCCCGACATTCCACAAGATCAAGTCAAATGGACGACAATGATCGAGAAGCTCAATCCGGACC TTCTGACACCACCCACTCCGAAAGAGCATTTCTATCCCTACACGCTCCCTTCGTATCGTGCAAAGCTACGACAGG AAGAGGAGCGAAGGCAACAACGTCAACAGGATACTCACAGGACGCCGACAAGGTCCTCAAGGCACGAACAATATC AAGAAGATGACGTCGAGTCCGACGCCGCGCCTCCAACATCCCGCTTAACCTCCTCTTCCCCACTCTTATTCCCTC GATCCTCACCCGCGAACCATGTCGACGAGGACATTCCTTCATCACCAGGTTCCGACTCTTTACAAGCCTCATGGG AACAAGGCATCCGAGACGCCATGGACCTCGTTACCAAACTCCTCGAACCCGAGTCCGTAAAGAGGATAACACCAG CCGCAGCCCTCGCCCACCCTTTCCTCCGCGAAGAAGATAAACTTGACGATGACTACTATGTTCCACACGTTTTCG GACGAGGTGTTTGTGGCAAGTACCATTTCTATGATGATGTGACGGAGCAGCCGGGGGTTATCATTAGGGAGAGGA GACGGAAGTGTGTCTGTAATTGCGGACTGGAAGAGTGCCAGGAGCAGGATGAGAATGCAGGGGAGTGGGTGAAGG TGAGGAAGCTGGTCTCCGCTGGCGAGGGGATAGCGATTGGGATGGAGCCATGCGAGTTCCACCAAGATTACCAAC TTTACGACGATGATGAGGACGATTGAAGGCATAAACCAACTGTATTCTTCTAGCATTGTACCTAACTTATTCTCG CTACTCAAGTAATCAATGCACGTTTCATTTACCCGCG |
| Length | 2437 |
Gene
| Sequence id | CopciAB_445594.T0 |
|---|---|
| Sequence |
>CopciAB_445594.T0 GGAAATTGGATCTCGTTCTGTCAACGGGCTTCTTTCTTAGCTCAATGTCTGTCACTTATTTTTCCCAACACTCTT CTAATCCGCTCGAGGCAAGGACTTCCGATGCCCGCAATCTTCATTACAACCGCGTCTACGCAGAGGCCAACGCCC AACTCGACGAAGTCTCTCCCTCGAGCGATCCGCTAGCCCGATATCCTCAAGACGAACTGCGCGAATACCACGAAC ATCTTGCAGAGGTCATTCCAGGGCACCCTCAGACAATGAGCCAAGCGCGGAAGAAAGAGACACGGATGGTGGAGC TCGACGAGGACGGTGACGGCGAAGAGGAGATGAGTGACGACGAGCTCATGATCATCCCCAAGGAGGAAGAGGACG TCTCCGTCGACGATGGAGAACAGGACAAGGAAGGCAACACCCCACAGCAACAGCAATTCGCCGAGTACGACGAGG ACATGGATGCAGAGGGCGAATGGGACGAGGACGTAGAGGTTGAAGAGCCCAAGGAGAAGGATTATTTCGCCCTGG ACGAATCTGAAGAAATGGGCTCGATAGACAAGAAAACCGAAGACGAAATTGCCGAAATCGAGAAGGAAATCGAAG AGTTTGAAGTGGCCCTTCCGACACTCGCTTCTGGGTACAAACTCCTCGACCGGTTGGGCACTGGGACGTTTTCTT CGGTCTACAAAGCACTCGACCTGCGGTATCACGAATACGACAACAGTCCATGGCTAGGACACCATCCGGAGGAGT CTTCAGCTCACTTCCAGTCCGTTCCGCAACCAGAGGGGTCGAGGGTATTCGTGGCTATCAAGAGGATTTACGTTA CAAGTGGCCCGGAGAGGATACGGAACGAACTGTTCATCATGGAGACGTGCAGGGGAGCAAGACACGTTTCGCAGC TCATCACCGCGTTCAGGCACCATGACCAGGTCGCCATCGTTATGCCTTACCATCGGAATGAGGATTTCAGGGTAA GTACCATGGAGCTTCTTCAATTGCGCTGAACTTATCCGAGGTGCAGGACTACTACGAGACGCTACCGATGGAAGG AATCAAATCCTATTTCCGATGTCTACTACGCGCATTACGCGATATCCACGCGAGGGGGATAATCCATCGCGACGT CAAACCAGCCAACTTTTTGTTTGATCCCAGCACCGGTCAGGGGACCCTGTGCGACTTTGGGTTGGCTCAGGTACG CCTGTTATACAGCGGCCTACCCGCATGACTGACAAGCACCACTGCAACAAGATCATGGACGACAAGCCTCCTGTG CTAGGCGCATGCCTGCACACGCAGCCCACGGAGCAATACCCCCACGGTCACATCCGGAAGAAACACCAGTACGAC GAGAATCTGGTGAAGGAGGGACAGATGGACGCGCGGAAGAAGAGTAAAATGCATGCGCATTCTGTTGGTATTCTT GACAAAGATACTCGGTACTGTTGCCTTTCCACCTGATGCGAAATATTGGTCGTGGAGACTGACCCGTGTCTCAGA ATGCCCATGAAAGCCAATCGTGCTGGTACTCGAGGATTCCGCGCACCTGAAGTCTTGCTCAAGTGCGGTCAGCAA TCCGGAGGTAAGGAGAGCCCGAGCTCCTTCCCCAGAGCTAGCTAACCCATTCGCGTATGGCAGCTGTGGATGTCT GGTCTGCGGGAATAATCCTCCTCTTCTTCCTCACTCGAAAGTTCCCAATATTCCAATCCAACGACGATATCGAGG CTCTCATGGAACTGGCTGTCATCTTCGGGCGGAAGAAAATGGAGAGCGTAGCCACACTACACGGTTGGTACTTTA TCTTATCTATGCCGCTCGTAATCTCACTCGTCCCCACAAAAAGCCCGCAAATTTGCAACCAACGTCCCCGACATT CCACAAGATCAAGTCAAATGGACGACAATGATCGAGAAGCTCAATCCGGACCTTCTGACACCACCCACTCCGAAA GAGCATTTCTATCCCTACACGCTCCCTTCGTATCGTGCAAAGCTACGACAGGAAGAGGAGCGAAGGCAACAACGT CAACAGGATACTCACAGGACGCCGACAAGGTCCTCAAGGCACGAACAATATCAAGAAGATGACGTCGAGTCCGAC GCCGCGCCTCCAACATCCCGCTTAACCTCCTCTTCCCCACTCTTATTCCCTCGATCCTCACCCGCGAACCATGTC GACGAGGACATTCCTTCATCACCAGGTTCCGACTCTTTACAAGCCTCATGGGAACAAGGCATCCGAGACGCCATG GACCTCGTTACCAAACTCCTCGAACCCGAGTCCGTAAAGAGGATAACACCAGCCGCAGCCCTCGCCCACCCTTTC CTCCGCGAAGAAGATAAACTTGACGATGACTACTATGTTCCACACGTTTTCGGACGAGGTGTTTGTGGCAAGTAC CATTTCTATGATGATGTGACGGAGCAGCCGGGGGTTATCATTAGGGAGAGGAGACGGAAGTGTGTCTGTAATTGC GGACTGGAAGAGTGCCAGGAGCAGGATGAGAATGCAGGGGAGTGGGTGAAGGTGAGGAAGCTGGTCTCCGCTGGC GAGGGGATAGCGATTGGGATGGAGCCATGCGAGTTCCACCAAGATTACCAACTTTACGACGATGATGAGGACGAT TGAAGGCATAAACCAACTGTATTCTTCTAGCATTGTACCTAACTTATTCTCGCTACTCAAGTAATCAATGCACGT TTCATTTACCCGCG |
| Length | 2714 |