CopciAB_449009
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_449009 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 449009 |
| Uniprot id | Functional description | GMC oxidoreductase | |
| Location | scaffold_3:982708..985545 | Strand | - |
| Gene length (nt) | 2838 | Transcript length (nt) | 2082 |
| CDS length (nt) | 1917 | Protein length (aa) | 638 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7266838 | 66.5 | 9.516E-285 | 877 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25944 | 59.8 | 1.15E-246 | 767 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114921 | 58.3 | 8.563E-236 | 735 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191146 | 58 | 1.979E-234 | 731 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_16977 | 50.5 | 9.351E-199 | 628 |
| Lentinula edodes NBRC 111202 | Lenedo1_1061307 | 49.4 | 9.334E-189 | 599 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_117168 | 49.4 | 9.258E-188 | 596 |
| Flammulina velutipes | Flave_chr09AA00940 | 52.8 | 2.224E-186 | 592 |
| Auricularia subglabra | Aurde3_1_1332730 | 48.6 | 6.666E-179 | 571 |
| Grifola frondosa | Grifr_OBZ72180 | 50.7 | 2.891E-172 | 551 |
| Lentinula edodes B17 | Lened_B_1_1_12376 | 46.8 | 1.489E-83 | 291 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_449009.T0 |
| Description | GMC oxidoreductase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 64 | 371 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 484 | 625 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR012132 | Glucose-methanol-choline oxidoreductase |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0050660 | flavin adenine dinucleotide binding | MF |
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
KEGG
| KEGG Orthology |
|---|
| K00108 |
EggNOG
| COG category | Description |
|---|---|
| E | GMC oxidoreductase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
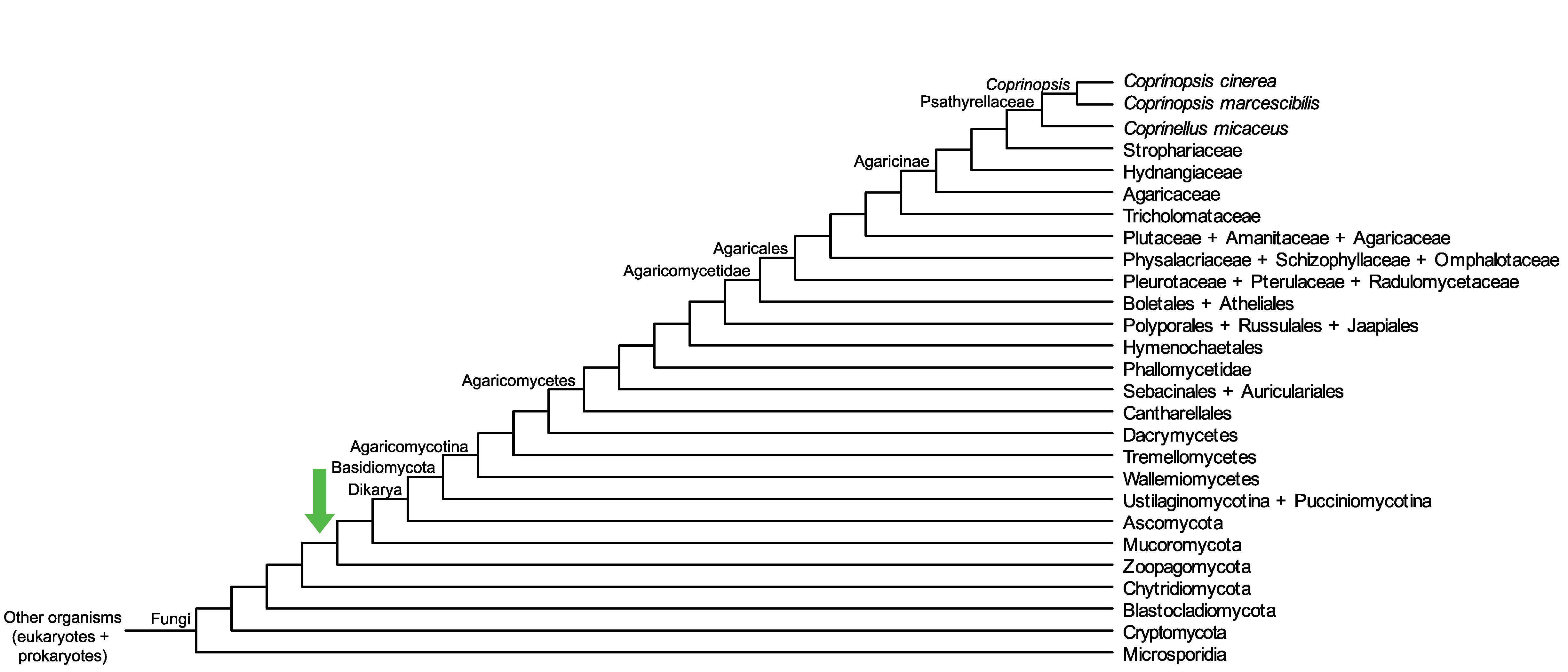
Conservation of CopciAB_449009 across fungi.
Arrow shows the origin of gene family containing CopciAB_449009.
Protein
| Sequence id | CopciAB_449009.T0 |
|---|---|
| Sequence |
>CopciAB_449009.T0 MSQASITFNNLSTRLGNNRFTWITAFAFALLLRQATKGRKPKLIADARRVAREVKEGELDFDEYDVVIIGGGTAG CVLAARLSEDPSLRVLLLEAGGSGVALRETRIPVAYSLLFHTKHVYQFFTEPQDFANGKKRFWPRAKMLGGCSSI NAQMAQWGSPGDFDRWGDIIGDDSWKWSNLKRYFNKFEKYVPHPDFPDVDVSVKGQEGPMKVGYFSEVSEGSKLF IKACQNVGIPFSPDFNTSKGTLGVNKIMTFIDETRTRVSSESAYFTPEVLSRPNLKVVVNATVTKILTERVGGEI RTVGVEFAKSKEGPRYRVRARKEVVLSAGAVHSPQILMLSGIGPSGELRKHHIPSVLDLPGVGDNLIDHPVVDLY FKNRYDDSPKHVKPSSLGDVVKFVTSTLKYFITRSGSMATNFGESAAFVRSDDRALFPESAYPEKLHDTTSSAIG PDLEIFTTPMAYKEHGDFTFPMHTFAIHVVLLRPRSKGTLRLKSASPWDAPAMNPTYLERPEDVHKLVRGVRLIT KIARQEPLVQRLDHSDKDPLLDSDTYLKSDKELEELVRERLETLYHPTSTCRMAPLEDGGVVDSRLRVYGVKGLR VCDASIFPEIVSGHTAAAVLATAEHLADIIKADLKEGH |
| Length | 638 |
Coding
| Sequence id | CopciAB_449009.T0 |
|---|---|
| Sequence |
>CopciAB_449009.T0 ATGTCCCAAGCAAGTATTACTTTCAACAACCTCTCAACAAGGCTCGGAAACAACAGGTTTACATGGATAACCGCG TTTGCGTTTGCGCTGTTGCTCCGACAGGCTACCAAGGGACGCAAACCAAAGCTCATTGCAGACGCGCGCCGGGTT GCTCGTGAGGTCAAAGAGGGGGAGCTTGACTTTGACGAGTACGACGTTGTTATCATTGGAGGAGGTACTGCTGGA TGCGTTCTCGCTGCCCGCCTTTCGGAGGATCCGTCTTTGCGAGTACTCTTGCTCGAGGCGGGTGGAAGTGGTGTA GCACTCCGGGAGACTCGGATTCCAGTTGCCTACTCGCTCTTGTTCCATACCAAGCATGTGTACCAATTCTTCACT GAGCCCCAGGACTTTGCCAATGGGAAGAAGAGGTTCTGGCCGAGAGCTAAGATGCTCGGGGGATGTTCTTCAATA AATGCTCAGATGGCACAATGGGGTTCACCAGGGGATTTCGATCGCTGGGGCGACATCATCGGCGACGATTCTTGG AAATGGAGCAATCTGAAACGATACTTCAACAAGTTTGAGAAATACGTTCCTCACCCTGACTTCCCTGACGTCGAT GTCAGCGTGAAAGGCCAGGAAGGCCCAATGAAAGTTGGTTATTTCAGCGAGGTGTCCGAAGGATCCAAGCTCTTC ATCAAGGCATGCCAGAATGTCGGGATCCCATTCAGTCCTGACTTCAATACCTCCAAAGGTACCCTTGGTGTGAAC AAGATCATGACCTTCATTGACGAGACCAGGACTCGTGTCTCCAGCGAGTCAGCCTACTTTACCCCCGAGGTCCTT TCTCGGCCTAATCTGAAGGTAGTGGTCAACGCCACTGTCACCAAGATCCTGACGGAGCGAGTGGGCGGAGAAATC AGAACCGTCGGTGTTGAATTTGCGAAGAGCAAGGAGGGGCCACGCTATCGCGTCAGGGCTAGGAAAGAGGTCGTC CTCTCAGCCGGTGCTGTCCACTCACCTCAGATTCTTATGCTTTCTGGAATTGGTCCCTCTGGAGAACTCCGTAAA CACCACATCCCTTCTGTTCTCGACCTTCCAGGTGTAGGAGACAACCTCATCGACCACCCCGTCGTCGACCTCTAC TTCAAGAACAGATACGACGACTCTCCGAAACACGTCAAGCCCAGCTCTCTCGGAGACGTCGTCAAGTTCGTGACT TCCACTCTCAAATACTTTATAACTCGGTCCGGATCCATGGCTACCAACTTTGGAGAGTCGGCCGCGTTCGTTCGA AGCGACGATCGTGCTTTGTTCCCAGAATCCGCGTACCCAGAAAAGCTTCATGATACGACCTCATCTGCCATCGGT CCCGACCTCGAGATCTTCACGACGCCGATGGCATACAAGGAACACGGCGACTTCACCTTCCCGATGCACACCTTC GCGATCCACGTTGTGCTTTTGAGACCGCGAAGCAAAGGTACTCTTCGTCTCAAGTCTGCTAGCCCTTGGGATGCG CCGGCTATGAATCCGACCTACCTCGAACGGCCGGAAGACGTCCACAAACTCGTCCGGGGCGTCCGTCTCATTACC AAGATCGCGCGTCAAGAGCCCCTCGTCCAACGCCTCGACCACTCGGATAAAGACCCCCTCCTCGACTCTGACACC TACCTCAAGAGCGACAAAGAACTCGAAGAACTCGTCCGCGAACGTCTCGAGACTTTGTACCACCCCACTTCGACT TGTAGGATGGCTCCGCTGGAAGACGGCGGTGTTGTGGATTCTCGTCTGCGTGTGTATGGTGTCAAGGGTCTCCGC GTTTGTGACGCATCGATCTTCCCCGAGATCGTGTCTGGACATACCGCTGCTGCAGTTCTTGCCACCGCTGAACAT CTCGCCGACATCATCAAGGCTGACTTGAAAGAGGGCCATTGA |
| Length | 1917 |
Transcript
| Sequence id | CopciAB_449009.T0 |
|---|---|
| Sequence |
>CopciAB_449009.T0 AAAGTTACTTTTTACGCACTCTCCCAGAACGACATTCATCTCGCCATGTCCCAAGCAAGTATTACTTTCAACAAC CTCTCAACAAGGCTCGGAAACAACAGGTTTACATGGATAACCGCGTTTGCGTTTGCGCTGTTGCTCCGACAGGCT ACCAAGGGACGCAAACCAAAGCTCATTGCAGACGCGCGCCGGGTTGCTCGTGAGGTCAAAGAGGGGGAGCTTGAC TTTGACGAGTACGACGTTGTTATCATTGGAGGAGGTACTGCTGGATGCGTTCTCGCTGCCCGCCTTTCGGAGGAT CCGTCTTTGCGAGTACTCTTGCTCGAGGCGGGTGGAAGTGGTGTAGCACTCCGGGAGACTCGGATTCCAGTTGCC TACTCGCTCTTGTTCCATACCAAGCATGTGTACCAATTCTTCACTGAGCCCCAGGACTTTGCCAATGGGAAGAAG AGGTTCTGGCCGAGAGCTAAGATGCTCGGGGGATGTTCTTCAATAAATGCTCAGATGGCACAATGGGGTTCACCA GGGGATTTCGATCGCTGGGGCGACATCATCGGCGACGATTCTTGGAAATGGAGCAATCTGAAACGATACTTCAAC AAGTTTGAGAAATACGTTCCTCACCCTGACTTCCCTGACGTCGATGTCAGCGTGAAAGGCCAGGAAGGCCCAATG AAAGTTGGTTATTTCAGCGAGGTGTCCGAAGGATCCAAGCTCTTCATCAAGGCATGCCAGAATGTCGGGATCCCA TTCAGTCCTGACTTCAATACCTCCAAAGGTACCCTTGGTGTGAACAAGATCATGACCTTCATTGACGAGACCAGG ACTCGTGTCTCCAGCGAGTCAGCCTACTTTACCCCCGAGGTCCTTTCTCGGCCTAATCTGAAGGTAGTGGTCAAC GCCACTGTCACCAAGATCCTGACGGAGCGAGTGGGCGGAGAAATCAGAACCGTCGGTGTTGAATTTGCGAAGAGC AAGGAGGGGCCACGCTATCGCGTCAGGGCTAGGAAAGAGGTCGTCCTCTCAGCCGGTGCTGTCCACTCACCTCAG ATTCTTATGCTTTCTGGAATTGGTCCCTCTGGAGAACTCCGTAAACACCACATCCCTTCTGTTCTCGACCTTCCA GGTGTAGGAGACAACCTCATCGACCACCCCGTCGTCGACCTCTACTTCAAGAACAGATACGACGACTCTCCGAAA CACGTCAAGCCCAGCTCTCTCGGAGACGTCGTCAAGTTCGTGACTTCCACTCTCAAATACTTTATAACTCGGTCC GGATCCATGGCTACCAACTTTGGAGAGTCGGCCGCGTTCGTTCGAAGCGACGATCGTGCTTTGTTCCCAGAATCC GCGTACCCAGAAAAGCTTCATGATACGACCTCATCTGCCATCGGTCCCGACCTCGAGATCTTCACGACGCCGATG GCATACAAGGAACACGGCGACTTCACCTTCCCGATGCACACCTTCGCGATCCACGTTGTGCTTTTGAGACCGCGA AGCAAAGGTACTCTTCGTCTCAAGTCTGCTAGCCCTTGGGATGCGCCGGCTATGAATCCGACCTACCTCGAACGG CCGGAAGACGTCCACAAACTCGTCCGGGGCGTCCGTCTCATTACCAAGATCGCGCGTCAAGAGCCCCTCGTCCAA CGCCTCGACCACTCGGATAAAGACCCCCTCCTCGACTCTGACACCTACCTCAAGAGCGACAAAGAACTCGAAGAA CTCGTCCGCGAACGTCTCGAGACTTTGTACCACCCCACTTCGACTTGTAGGATGGCTCCGCTGGAAGACGGCGGT GTTGTGGATTCTCGTCTGCGTGTGTATGGTGTCAAGGGTCTCCGCGTTTGTGACGCATCGATCTTCCCCGAGATC GTGTCTGGACATACCGCTGCTGCAGTTCTTGCCACCGCTGAACATCTCGCCGACATCATCAAGGCTGACTTGAAA GAGGGCCATTGAATGGACGGCCATAGGTAGATACAACAGGGGATATTCAGGTCGGACGGAGGAGGAATTAAATTA AGAGGTAGTACAGACAACACTGTATAACAACAATCTTTGATTCTGGATATGCATAGG |
| Length | 2082 |
Gene
| Sequence id | CopciAB_449009.T0 |
|---|---|
| Sequence |
>CopciAB_449009.T0 AAAGTTACTTTTTACGCACTCTCCCAGAACGACATTCATCTCGCCATGTCCCAAGCAAGTATTACTTTCAACAAC CTCTCAACAAGGCTCGGAAACAACAGGTTTACATGGATAACCGCGTTTGCGTTTGCGCTGTTGCTCCGACAGGCT ACCAAGGGACGCAAACCAAAGCTCATTGCAGACGCGCGCCGGGTTGCTCGTGAGGTCAAAGAGGGGGAGCTTGAC TTTGACGAGTACGACGTTGTTATCATTGGAGGAGGTACGTGACAAGGCAGAGAATACCCCCAGGGCTTACCTCTG ACCTACCTTCCTCAGGTACTGCTGGATGCGTTCTCGCTGCCCGCCTTTCGGAGGATCCGTCTTTGCGAGTACTCT TGCTCGAGGCGGGTGGAAGGTACACTTTGTGTATTCTCGCTTCCAGTATTTTGACTTCCGGTTCAATAGTGGTGT AGCACTCCGGGAGACTCGGATTCCAGTTGCCTACTCGCTCTTGTTCCATACCAAGCATGTGTACCAATTCTTCAC TGAGCCCCAGGACTTTGCCAATGGGAAGAAGAGGTTCTGGCCGAGAGGTAATCTTACTTTGTCGCTTGCCTTAAG TATTTGCCCACACTCACCTGGAATCTAGCTAAGATGCTCGGGGGATGTTCTTCAATAAATGCTCAGATGTACGTC TCGGAGCCCCTAATTCCCATCGATTCGCCAACCTCATGCAATCAGGGCACAATGGGGTTCACCAGGGGATTTCGA TCGCTGGGGCGACATCATCGGCGACGATTCTTGGAAATGGAGCAATCTGAAACGGTATGGGATGAGGGGCACCCA TGTGTGTATCCTGCTAACTGCGATGCTGTGCCAGATACTTCAACAAGTTTGAGAAATACGTTCCTCACCCTGACT TCCCTGACGTCGATGTCAGCGTGAAAGGCCAGGAAGGCCCAATGAAAGTTGGTTATTTCAGCGAGGTGTCCGAAG GATCCAAGCTCTTCATCAAGGCATGCCAGAATGTCGGGATCCCATTCAGTCCTGACTTCAATACCTCCAAAGGTA CCCTTGGTGTGAACAAGGTATGTTGTTCATTAATTTTACTAGGGTTCGTCAAGTTGACTTTTGTAGATCAGTGTG TACCCTCAGCTCTTAGACGGTCCCCAATTTCTGACATATTTCCTCAGTGACCTTCATTGACGAGACCAGGACTCG TGTCTCCAGCGAGTCAGCCTACTTTACCCCCGAGGTCCTTTCTCGGCCTAATCTGAAGGTAGTGGTCAACGCCAC TGTCACCAAGATCCTGACGGAGCGAGTGGGCGGAGAAATCAGAACCGTCGGTGTTGAATTTGCGAAGAGCAAGGA GGGGCCACGCTATCGCGTCAGGGCTAGGAAAGAGGTCGTCCTCTCGTGCGTCCCTTCCCCCTTCTATTCAAGAAC AACTCATGACCGTAGTTACAACGCAGAGCCGGTGCTGTCCACTCACCTCAGGTCGGCCTTTTCCCCGACCGTTCG TTCCCTCTTACTGATATAGAATGTAGATTCTTATGCTTTCTGGAATTGGTCCCTCTGGAGAACTCCGTAAACACC ACATCCCTTCTGTTCTCGACCTTCCAGGTGTAGGAGACAACCTCATCGACCACCCCGTCGTCGACCTCTACTTCA AGAACAGATACGACGACTCTCCGAAACACGTCAAGCCCAGCTCTCTCGGAGACGTCGTCAAGTTCGTGACTTCCA CTCTCAAATACTTTATAACTCGGTCCGGATCCATGGCTACCAACGTCAGTCCTTGTGCACATGTCTGCCGTCCAC TTCGTGCTAACAGTCCTCTGTCGTTTCTAGTTTGGAGAGTCGGCCGCGTTCGTTCGAAGCGACGATCGTGCTTTG TTCCCAGAATCCGCGTACCCAGAAAAGCTTCATGATACGACCTCATCTGCCATCGGTCCCGACCTCGAGATCTTC ACGACGCCGATGGCATACAAGGTACAGATATATCTGCTGGTACGCGTTCCATGATGTCGACGCACTGTACTTCCA GGAACACGGCGACTTCACCTTCCCGATGCACACCTTCGCGATCCACGTTGTGCTTTTGAGGTACTTCACAATCAA ATGCCCTCCGATTGCTTTCGTTAACATTCACATTCCCTTAGACCGCGAAGCAAAGGTACTCTTCGTCTCAAGTCT GCTAGCCCTTGGGATGCGCCGGCTATGAATCCGACGTACGAATCTATTCTTCATCACCATTTCGCCATCTGACAC CTTTTTCTAGCTACCTCGAACGGCCGGAAGACGTCCACAAACTCGTCCGGGGCGTCCGTCTCATTACCAAGATCG CGCGTCAAGAGCCCCTCGTCCAACGCCTCGACCACTCGGATAAAGACCCCCTCCTCGACTCTGACACCTACCTCA AGAGCGACAAAGAACTCGAAGAACTCGTCCGCGAACGTCTCGAGACTTTGTACCACCCCACTTCGACTTGTAGGA TGGCTCCGCTGGAAGACGGCGGTGTTGTGGATTCTCGTCTGCGTGTGTATGGTGTCAAGGGTCTCCGCGTTTGTG ACGCATCGATCTTCCCCGAGATCGTGTCTGGACATACCGTAAGTTTGGCTTTTGGTTTGCAGCGTGGCTTTTGAC GCTGAAGTGAATCATGAACAGGCTGCTGCAGTTCTTGCCACCGCTGAACATCTCGCCGACATCATCAAGGCTGAC TTGAAAGAGGGCCATTGAATGGACGGCCATAGGTAGATACAACAGGGGATATTCAGGTCGGACGGAGGAGGAATT AAATTAAGAGGTAGTACAGACAACACTGTATAACAACAATCTTTGATTCTGGATATGCATAGG |
| Length | 2838 |