CopciAB_449440
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_449440 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | HRR25 | Synonyms | 449440 |
| Uniprot id | Functional description | Belongs to the protein kinase superfamily | |
| Location | scaffold_1:1331445..1334185 | Strand | + |
| Gene length (nt) | 2741 | Transcript length (nt) | 2229 |
| CDS length (nt) | 1098 | Protein length (aa) | 365 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | ck-1a |
| Aspergillus nidulans | AN4563_ckiA |
| Schizosaccharomyces pombe | hhp1 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264740 | 93.7 | 1.603E-230 | 705 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28600 | 93.3 | 5.284E-227 | 695 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1111729 | 92.5 | 4.626E-225 | 689 |
| Pleurotus ostreatus PC9 | PleosPC9_1_112679 | 92.5 | 4.528E-225 | 689 |
| Flammulina velutipes | Flave_chr08AA00386 | 92.3 | 1.107E-224 | 688 |
| Schizophyllum commune H4-8 | Schco3_2172656 | 92.2 | 6.716E-224 | 686 |
| Lentinula edodes B17 | Lened_B_1_1_16260 | 90.7 | 1.04E-223 | 685 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_342 | 90.7 | 1.305E-223 | 685 |
| Grifola frondosa | Grifr_OBZ78915 | 89.5 | 1.588E-220 | 676 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_62578 | 95.4 | 4.11E-216 | 663 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_31660 | 95.4 | 4.282E-216 | 663 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1437732 | 95.4 | 3.317E-213 | 655 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_37853 | 93.6 | 3.629E-211 | 649 |
| Lentinula edodes NBRC 111202 | Lenedo1_1032047 | 93.7 | 3.954E-211 | 649 |
| Auricularia subglabra | Aurde3_1_1023935 | 89.1 | 1.035E-196 | 608 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | HRR25 |
|---|---|
| Protein id | CopciAB_449440.T0 |
| Description | Belongs to the protein kinase superfamily |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14125 | STKc_CK1_delta_epsilon | - | 15 | 289 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 16 | 234 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K14758 |
EggNOG
| COG category | Description |
|---|---|
| T | Belongs to the protein kinase superfamily |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
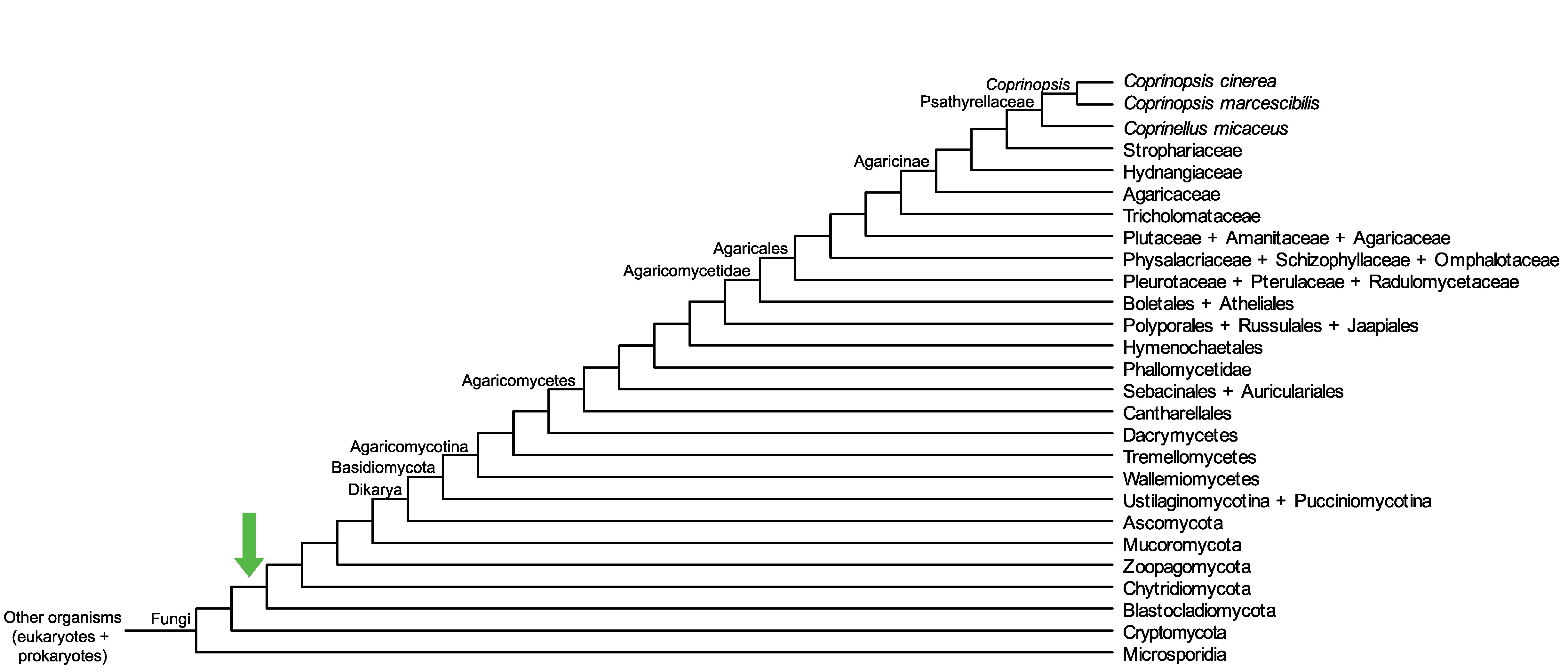
Conservation of CopciAB_449440 across fungi.
Arrow shows the origin of gene family containing CopciAB_449440.
Protein
| Sequence id | CopciAB_449440.T0 |
|---|---|
| Sequence |
>CopciAB_449440.T0 MAESAHRLDLRVGGKYRLGKKIGSGSFGDIYLGINIISGEEVAIKLESVKAKHPQLEYESKVYKTLAGGVGVPFV RWFGTECDYNAMVLDLLGPSLEDLFNFCNRKFSLKTVLLLADQLISRIEYIHSRNFIHRDIKPDNFLMGIGKRGN QVNVIDFGLAKKFRDPKTHLHIPYRENKNLTGTARYTSINTHLGVEQARRDDLESLAYVLMYFLRGALPWQGLKA ATKKQKYDRIMEKKMTTPTDLLCRGFPNEFGIFLNYTRALRFDDKPDYSYLRKLFRDLFVREGYQYDYVFDWSVQ RGAQEDQAASSSKVPVGRRKVVQEADEDHRASDRMLRSHTRQAQQAGGSGMPQARAGRGRDAEVW |
| Length | 365 |
Coding
| Sequence id | CopciAB_449440.T0 |
|---|---|
| Sequence |
>CopciAB_449440.T0 ATGGCCGAATCTGCGCACAGACTGGATCTTAGAGTAGGCGGCAAATACAGACTTGGGAAAAAGATCGGTTCAGGG TCATTTGGCGATATCTACCTTGGTATTAACATCATCTCTGGTGAAGAGGTCGCCATCAAGCTCGAATCAGTCAAG GCCAAGCACCCCCAGCTCGAGTATGAGTCCAAGGTCTACAAGACCCTCGCCGGCGGTGTTGGCGTCCCCTTTGTG AGGTGGTTTGGAACCGAGTGTGACTACAACGCCATGGTTCTCGACCTCCTCGGACCCTCTCTCGAGGATCTCTTC AACTTCTGCAACCGCAAGTTCAGCCTCAAGACCGTTCTTCTCCTCGCTGATCAACTGATCTCTCGCATCGAGTAC ATCCACTCTCGCAACTTCATCCACCGTGACATCAAGCCCGACAACTTCTTGATGGGCATCGGCAAACGTGGCAAC CAGGTCAACGTCATTGACTTTGGTCTCGCGAAGAAATTCCGCGACCCAAAGACACATCTCCATATTCCTTACCGA GAGAACAAGAATTTGACCGGTACCGCCCGCTACACTTCCATCAACACCCACTTGGGTGTCGAGCAAGCCCGTCGT GACGACCTCGAATCCCTCGCCTATGTCCTCATGTACTTCCTCCGCGGCGCTCTGCCCTGGCAAGGCCTCAAGGCT GCTACCAAGAAGCAGAAGTACGACCGTATCATGGAGAAGAAGATGACCACCCCCACCGACCTTCTCTGCCGTGGA TTCCCCAACGAGTTTGGCATTTTCTTGAACTATACCCGTGCCCTTCGCTTCGATGACAAGCCCGACTATTCGTAC CTTCGCAAGCTCTTCCGCGACCTCTTCGTTCGCGAAGGTTATCAGTACGATTACGTTTTCGACTGGAGTGTCCAG CGTGGAGCTCAGGAGGACCAAGCTGCCTCCAGCTCGAAGGTTCCAGTCGGCCGTCGCAAGGTCGTTCAGGAGGCT GACGAGGACCATCGTGCAAGCGACAGAATGCTTCGCTCCCACACCAGGCAGGCTCAGCAGGCCGGTGGATCCGGA ATGCCTCAGGCCAGGGCTGGCCGGGGTCGTGATGCTGAAGTCTGGTAA |
| Length | 1098 |
Transcript
| Sequence id | CopciAB_449440.T0 |
|---|---|
| Sequence |
>CopciAB_449440.T0 ACTTTCTTCCAAACAACCATCTATCTATCCTCTTTATCCCTTTTATCTCTCTCTCCCTCAGTAATCGATGGCCGA ATCTGCGCACAGACTGGATCTTAGAGTAGGCGGCAAATACAGACTTGGGAAAAAGATCGGTTCAGGGTCATTTGG CGATATCTACCTTGGTATTAACATCATCTCTGGTGAAGAGGTCGCCATCAAGCTCGAATCAGTCAAGGCCAAGCA CCCCCAGCTCGAGTATGAGTCCAAGGTCTACAAGACCCTCGCCGGCGGTGTTGGCGTCCCCTTTGTGAGGTGGTT TGGAACCGAGTGTGACTACAACGCCATGGTTCTCGACCTCCTCGGACCCTCTCTCGAGGATCTCTTCAACTTCTG CAACCGCAAGTTCAGCCTCAAGACCGTTCTTCTCCTCGCTGATCAACTGATCTCTCGCATCGAGTACATCCACTC TCGCAACTTCATCCACCGTGACATCAAGCCCGACAACTTCTTGATGGGCATCGGCAAACGTGGCAACCAGGTCAA CGTCATTGACTTTGGTCTCGCGAAGAAATTCCGCGACCCAAAGACACATCTCCATATTCCTTACCGAGAGAACAA GAATTTGACCGGTACCGCCCGCTACACTTCCATCAACACCCACTTGGGTGTCGAGCAAGCCCGTCGTGACGACCT CGAATCCCTCGCCTATGTCCTCATGTACTTCCTCCGCGGCGCTCTGCCCTGGCAAGGCCTCAAGGCTGCTACCAA GAAGCAGAAGTACGACCGTATCATGGAGAAGAAGATGACCACCCCCACCGACCTTCTCTGCCGTGGATTCCCCAA CGAGTTTGGCATTTTCTTGAACTATACCCGTGCCCTTCGCTTCGATGACAAGCCCGACTATTCGTACCTTCGCAA GCTCTTCCGCGACCTCTTCGTTCGCGAAGGTTATCAGTACGATTACGTTTTCGACTGGAGTGTCCAGCGTGGAGC TCAGGAGGACCAAGCTGCCTCCAGCTCGAAGGTTCCAGTCGGCCGTCGCAAGGTCGTTCAGGAGGCTGACGAGGA CCATCGTGCAAGCGACAGAATGCTTCGCTCCCACACCAGGCAGGCTCAGCAGGCCGGTGGATCCGGAATGCCTCA GGCCAGGGCTGGCCGGGGTCGTGATGCTGAAGTCTGGTAAACGGGCTTCTTAGTTTGGTTTCTGGCCTGTTTTCT CTATAGTGCGTCCCGAGAGTGTCGAGCGAATGGGGGGTGGAGGGCAGAAGCAGGTATCGCTCGGCAGTGACTGAT GATCAAGGACGTCGTATGGTTGGGTGGAGACGCGTCCCTGGGATCGTCAGGAGTATGTGGATGCGCGGAGGAGGT TGTAGAAGTTACGCGAGTGTTGGCGGGGGTTGTTGGATGGTTTCGTTTGGGCGTTTCCCCCAACTTGTTGCATAG ATCTGGGTTCACGGACAGCATCAGTTGACGCCCGGCGATTCCACCCCTATCATACCAAGTATTCCCTTCAAGAAG AAGCCTGGACAACCTCTCAACTCAATCGCAAGCTTCCCATGCCCAAGGTGTATTCGACCCTAAACATTATTCTGG CTTCATTGCCTGTTTACATCGCTTCTTTCATCTGAAACGGACGCTTTTTCTTTTCACGTCAGTCTCTTGTTCTTC GGAACAAGGGCTGGCCGCCTCACCCCCAACCCCTTGTTATCTATTATCATAATCCAAATCCTCACCCCCTGCGCC CTTTCAGCCTAACGTTAGCCCATTTCCCCCTTGTACACCTTCCCGTTCACTTGACCTGGTTTCCCTTTTTTTCAT CGGTAGTACACGATCCCCCATCACTCGTATTATTGACATTCATTGGCTTCAAGAATTTCCCCTTCCCCCACCATT CCTTTCTTCCCCCTTTCTGTTTCTGTATGGGTTTTAGGTCTTCACTTTATCCTTCCGTCCTCCACCTTTCTCTTC CCCAATGAGAAAGGGCGAGCGGGACGGAAGAGGGCGCCGAGTGTAGAAGGAGAGGTTACGTCGAGTGCTGGACTC GCGTGAGGAGAGTTGACGAACAGTTGTCGGTGTGGACAATAACAAGTTTCGTAGTAGACATGCGTCGTATGATCA CCCCTCGTTTCCACCCCTTTCGGTCTCTCCGAATCCCTCACCCCCTTCCGAATCGATGTCTACTGTTGCCTTCAA TGTATGACTATTGTGACGAATGTTGTTAATATGACCAGACACTTGTGTTCCGCG |
| Length | 2229 |
Gene
| Sequence id | CopciAB_449440.T0 |
|---|---|
| Sequence |
>CopciAB_449440.T0 ACTTTCTTCCAAACAACCATCTATCTATCCTCTTTATCCCTTTTATCTGTGAGTAGCCCTCTTTTTCTTTTCGTC CCCTCGTCGTCGTCGTCGCCCAGCGGGTGCCACACAGCGCCTGCCCTGCAACCGCACAGCTGTCCCATTCAGCCA TTGTACCCTTCCTCCTTTTCTCGCCCCGCCCGTCCTTGTTCCCAGAACAATGAGGACTCGGTCAAGCGACAACGA CAGGCGTCACCCCTGGTTAGCCACCGCAACGAGGCCGGCCTGTTGTCTGGGATCCACATGCCCCCAGCCCTGTTA TATATAAAGCCCATCATGCACTCCTTCAATTTTTTGCCTTCCTTCCCCCACTTTCTCTCTTCACCCTTACTTACA CCGGTCTCTAGCTCTCTCCCTCAGTAATCGATGGCCGAATCTGCGCACAGACTGGATCTTAGAGTAGGCGGCAAA TACAGACTTGGGAAAAAGATCGGTTCAGGGTCATTTGGTGCGTCGTCTCCGTCCGCAGGCGTCATGAATGATGTT ACTCACACTTGCATGCAGGCGATATCTACCTTGGTATTAACATCATCTCTGGTGAAGAGGTCGCCATCAAGCTCG AATCAGTCAAGGCCAAGCACCCCCAGCTCGAGTATGAGTCCAAGGTCTACAAGACCCTCGCCGGCGGTGTTGGCG TCCCCTTTGTGAGGTGGTTTGGAACCGAGTGTGACTACAACGCCATGGTTCTCGACCTCCTCGGACCCTCTCTCG AGGATCTCTTCAACTTCTGCAACCGCAAGTTCAGCCTCAAGACCGTTCTTCTCCTCGCTGATCAACTGGTGAGTC CGGGTCTCCCCCCTATATCCTTTTCATGCTCCGTTTGCTGATGTGGCATGCAGATCTCTCGCATCGAGTACATCC ACTCTCGCAACTTCATCCACCGTGACATCAAGCCCGACAACTTCTTGATGGGCATCGGCAAACGTGGCAACCAGG TCAACGTCATTGACTTTGGTCTCGCGAAGAAATTCCGCGACCCAAAGACACATCTCCATATTCCTTACCGAGAGA ACAAGAATTTGACCGGTACCGCCCGCTACACTTCCATCAACACCCACTTGGGTGTCGAGCAAGCCCGTCGTGACG ACCTCGAATCCCTCGCCTATGTCCTCATGTACTTCCTCCGCGGCGCTCTGCCCTGGCAAGGCCTCAAGGCTGCTA CCAAGAAGCAGAAGTACGACCGTATCATGGAGAAGAAGATGACCACCCCCACCGACCTTCTCTGCCGTGGATTCC CCAACGAGTTTGGCATTTTCTTGAACTATACCCGTGCCCTTCGCTTCGATGACAAGCCCGACTATTCGTACCTTC GCAAGCTCTTCCGCGACCTCTTCGTTCGCGAAGGTTATCAGTACGATTACGTTTTCGACTGGAGTGTCCAGCGTG GAGCTCAGGAGGACCAAGCTGCCTCCAGCTCGAAGGTTCCAGTCGGCCGTCGCAAGGTCGTTCAGGAGGCTGACG AGGACCATCGTGCAAGCGACAGAATGTAAGCATGACTTTACCCTTACTCCAGTCACCCAGATCGCGCTAACAATT GTGAATAGGCTTCGCTCCCACACCAGGCAGGCTCAGCAGGCCGGTGGATCCGGAATGCCTCAGGCCAGGGCTGGC CGGGGTCGTGATGCTGAAGTCTGGTAAACGGGCTTCTTAGTTTGGTTTCTGGCCTGTTTTCTCTATAGTGCGTCC CGAGAGTGTCGAGCGAATGGGGGGTGGAGGGCAGAAGCAGGTATCGCTCGGCAGTGACTGATGATCAAGGACGTC GTATGGTTGGGTGGAGACGCGTCCCTGGGATCGTCAGGAGTATGTGGATGCGCGGAGGAGGTTGTAGAAGTTACG CGAGTGTTGGCGGGGGTTGTTGGATGGTTTCGTTTGGGCGTTTCCCCCAACTTGTTGCATAGATCTGGGTTCACG GACAGCATCAGTTGACGCCCGGCGATTCCACCCCTATCATACCAAGTATTCCCTTCAAGAAGAAGCCTGGACAAC CTCTCAACTCAATCGCAAGCTTCCCATGCCCAAGGTGTATTCGACCCTAAACATTATTCTGGCTTCATTGCCTGT TTACATCGCTTCTTTCATCTGAAACGGACGCTTTTTCTTTTCACGTCAGTCTCTTGTTCTTCGGAACAAGGGCTG GCCGCCTCACCCCCAACCCCTTGTTATCTATTATCATAATCCAAATCCTCACCCCCTGCGCCCTTTCAGCCTAAC GTTAGCCCATTTCCCCCTTGTACACCTTCCCGTTCACTTGACCTGGTTTCCCTTTTTTTCATCGGTAGTACACGA TCCCCCATCACTCGTATTATTGACATTCATTGGCTTCAAGAATTTCCCCTTCCCCCACCATTCCTTTCTTCCCCC TTTCTGTTTCTGTATGGGTTTTAGGTCTTCACTTTATCCTTCCGTCCTCCACCTTTCTCTTCCCCAATGAGAAAG GGCGAGCGGGACGGAAGAGGGCGCCGAGTGTAGAAGGAGAGGTTACGTCGAGTGCTGGACTCGCGTGAGGAGAGT TGACGAACAGTTGTCGGTGTGGACAATAACAAGTTTCGTAGTAGACATGCGTCGTATGATCACCCCTCGTTTCCA CCCCTTTCGGTCTCTCCGAATCCCTCACCCCCTTCCGAATCGATGTCTACTGTTGCCTTCAATGTATGACTATTG TGACGAATGTTGTTAATATGACCAGACACTTGTGTTCCGCG |
| Length | 2741 |