CopciAB_450167
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_450167 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | chk1 | Synonyms | 450167 |
| Uniprot id | Functional description | Belongs to the protein kinase superfamily | |
| Location | scaffold_1:4033411..4035820 | Strand | + |
| Gene length (nt) | 2410 | Transcript length (nt) | 1633 |
| CDS length (nt) | 1377 | Protein length (aa) | 458 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Ustilago maydis | CHK1 |
| Schizosaccharomyces pombe | chk1 |
| Saccharomyces cerevisiae | YBR274W_CHK1 |
| Aspergillus nidulans | AN5494_chkA |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB27406 | 69.4 | 4.024E-215 | 666 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1094565 | 65.7 | 9.27E-211 | 653 |
| Pleurotus ostreatus PC9 | PleosPC9_1_113089 | 65.7 | 9.074E-211 | 653 |
| Agrocybe aegerita | Agrae_CAA7269682 | 65.5 | 3.271E-207 | 643 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1433090 | 64.4 | 9.119E-204 | 633 |
| Flammulina velutipes | Flave_chr09AA00880 | 63.5 | 9.123E-203 | 630 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_122240 | 63.7 | 4.685E-199 | 619 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_132921 | 63.2 | 5.33E-198 | 616 |
| Lentinula edodes NBRC 111202 | Lenedo1_1123749 | 57 | 1.656E-187 | 586 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_21078 | 56.8 | 8.082E-187 | 584 |
| Grifola frondosa | Grifr_OBZ78002 | 61.2 | 1.656E-186 | 583 |
| Auricularia subglabra | Aurde3_1_1248264 | 56.1 | 1.498E-166 | 526 |
| Schizophyllum commune H4-8 | Schco3_2482682 | 61.5 | 1.815E-150 | 479 |
| Lentinula edodes B17 | Lened_B_1_1_16822 | 55 | 5.912E-131 | 422 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_69436 | 57.3 | 1.282E-122 | 398 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | chk1 |
|---|---|
| Protein id | CopciAB_450167.T0 |
| Description | Belongs to the protein kinase superfamily |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 10 | 278 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K02216 |
EggNOG
| COG category | Description |
|---|---|
| D | Belongs to the protein kinase superfamily |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
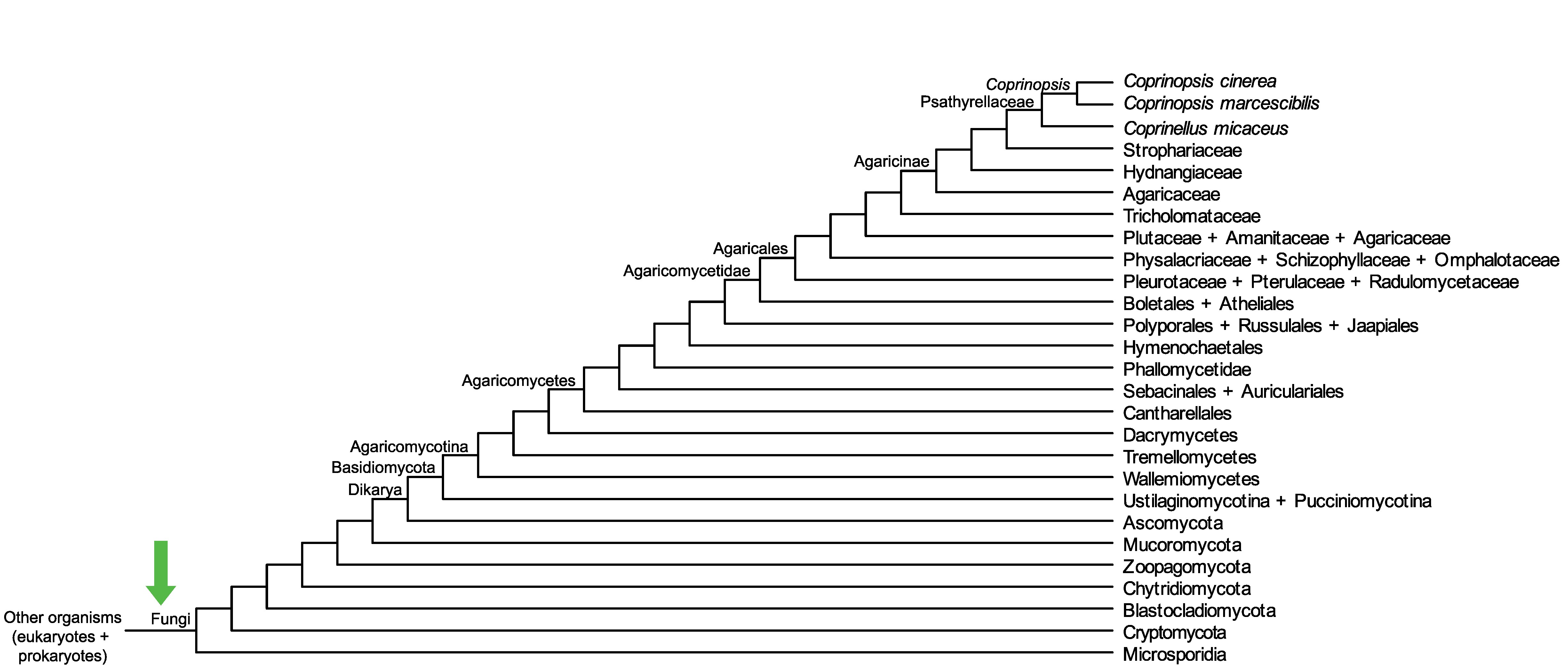
Conservation of CopciAB_450167 across fungi.
Arrow shows the origin of gene family containing CopciAB_450167.
Protein
| Sequence id | CopciAB_450167.T0 |
|---|---|
| Sequence |
>CopciAB_450167.T0 MPEFPKVTGYTLVDRIGGGGFSSVYKAIDFKNYRAAACKVIIITEETTDKERKTLDKEMRIHSALKHENVLEFLN AVVVELKHKHAYHPGYYMLMELACGGDLFDKIAPDKGVSDDLAHLYFNQLIAGMDYIHQQGVCHRDLKPENLLLD VAGTLKISDFGLSSVYKLKDSGRTRSLTERCGSLPYVAPELNSNAPYEAEPIDVWGCGVILFTLLVGNTPWDEPT RHSPEFCRYLDGTIFDEDPWNRISETPLSLLQGMLNVNPAERMTLSDVYQHPWCLRPSQLARADPGTLAEMLTQG LREDGDLNVAAPDFGEAMDVDEEDDPQRYAGTASQFTQKLMLFSQTQSGPRYVPNLTRFYASAPPTVLMPLIQET LEGMNVKCKLDPRSKMVLLLRIGGYDKRKLKFRGWVNVESFSKHGYEGSFCGMARDEGSPISWRQLWKALIKSPL VSPHVLFK |
| Length | 458 |
Coding
| Sequence id | CopciAB_450167.T0 |
|---|---|
| Sequence |
>CopciAB_450167.T0 ATGCCCGAATTCCCAAAGGTTACAGGGTACACGCTAGTGGACAGAATTGGAGGCGGCGGGTTCTCCAGCGTTTAT AAAGCTATAGACTTCAAGAACTACCGTGCAGCAGCATGTAAAGTGATCATCATTACTGAGGAAACCACAGATAAA GAGAGAAAGACACTCGACAAGGAAATGCGGATCCATTCGGCGCTGAAGCACGAAAACGTGCTGGAATTTCTGAAC GCTGTGGTCGTCGAGCTCAAGCACAAACATGCCTATCATCCGGGATATTATATGCTAATGGAGCTGGCTTGTGGT GGAGATCTGTTCGATAAGATCGCGCCCGACAAGGGCGTCAGTGACGACCTGGCACATCTGTATTTTAACCAGTTG ATAGCCGGAATGGACTATATCCACCAACAAGGCGTGTGTCATCGCGACCTTAAGCCGGAGAATCTACTACTGGAC GTCGCTGGGACGCTCAAGATCTCCGATTTTGGTTTATCTTCAGTGTACAAACTGAAAGACTCGGGTCGGACGCGA TCTCTGACCGAACGCTGTGGAAGCCTCCCGTATGTTGCACCAGAGCTGAACAGCAACGCGCCGTACGAAGCGGAG CCAATCGATGTCTGGGGATGTGGGGTCATCCTCTTTACTCTGTTGGTCGGTAATACCCCATGGGACGAGCCCACC CGACACAGCCCAGAATTCTGCCGCTATCTGGATGGGACCATCTTCGACGAAGATCCATGGAATCGGATCTCTGAG ACCCCGCTGTCTCTCCTTCAGGGTATGCTGAACGTGAACCCAGCTGAGCGTATGACGCTGTCTGACGTGTATCAG CATCCATGGTGTTTACGACCGAGCCAATTGGCACGAGCAGACCCAGGTACCCTGGCGGAGATGCTGACGCAGGGG CTACGCGAGGACGGCGATCTAAATGTAGCAGCCCCTGATTTCGGAGAAGCTATGGATGTCGACGAAGAGGACGAT CCCCAGCGTTATGCTGGAACAGCCTCTCAATTCACGCAGAAACTCATGCTTTTCTCGCAAACGCAATCGGGCCCT CGATATGTGCCGAACCTGACTCGATTCTACGCTTCTGCGCCGCCAACGGTATTGATGCCCCTGATCCAAGAAACA CTGGAGGGAATGAACGTCAAATGTAAACTTGACCCTAGATCTAAGATGGTGCTGCTGTTGAGGATCGGAGGGTAC GACAAGCGCAAGCTCAAGTTCCGAGGGTGGGTGAACGTGGAGAGCTTCAGCAAACATGGGTACGAGGGGAGCTTC TGCGGCATGGCGAGAGATGAGGGGAGCCCGATTTCGTGGCGGCAGCTGTGGAAGGCCCTCATCAAGAGTCCCCTG GTCAGCCCCCACGTCCTATTCAAATAG |
| Length | 1377 |
Transcript
| Sequence id | CopciAB_450167.T0 |
|---|---|
| Sequence |
>CopciAB_450167.T0 AGCGTTTCTTTCTTCTACCACCACAACGGCGCCAAAATGCCCGAATTCCCAAAGGTTACAGGGTACACGCTAGTG GACAGAATTGGAGGCGGCGGGTTCTCCAGCGTTTATAAAGCTATAGACTTCAAGAACTACCGTGCAGCAGCATGT AAAGTGATCATCATTACTGAGGAAACCACAGATAAAGAGAGAAAGACACTCGACAAGGAAATGCGGATCCATTCG GCGCTGAAGCACGAAAACGTGCTGGAATTTCTGAACGCTGTGGTCGTCGAGCTCAAGCACAAACATGCCTATCAT CCGGGATATTATATGCTAATGGAGCTGGCTTGTGGTGGAGATCTGTTCGATAAGATCGCGCCCGACAAGGGCGTC AGTGACGACCTGGCACATCTGTATTTTAACCAGTTGATAGCCGGAATGGACTATATCCACCAACAAGGCGTGTGT CATCGCGACCTTAAGCCGGAGAATCTACTACTGGACGTCGCTGGGACGCTCAAGATCTCCGATTTTGGTTTATCT TCAGTGTACAAACTGAAAGACTCGGGTCGGACGCGATCTCTGACCGAACGCTGTGGAAGCCTCCCGTATGTTGCA CCAGAGCTGAACAGCAACGCGCCGTACGAAGCGGAGCCAATCGATGTCTGGGGATGTGGGGTCATCCTCTTTACT CTGTTGGTCGGTAATACCCCATGGGACGAGCCCACCCGACACAGCCCAGAATTCTGCCGCTATCTGGATGGGACC ATCTTCGACGAAGATCCATGGAATCGGATCTCTGAGACCCCGCTGTCTCTCCTTCAGGGTATGCTGAACGTGAAC CCAGCTGAGCGTATGACGCTGTCTGACGTGTATCAGCATCCATGGTGTTTACGACCGAGCCAATTGGCACGAGCA GACCCAGGTACCCTGGCGGAGATGCTGACGCAGGGGCTACGCGAGGACGGCGATCTAAATGTAGCAGCCCCTGAT TTCGGAGAAGCTATGGATGTCGACGAAGAGGACGATCCCCAGCGTTATGCTGGAACAGCCTCTCAATTCACGCAG AAACTCATGCTTTTCTCGCAAACGCAATCGGGCCCTCGATATGTGCCGAACCTGACTCGATTCTACGCTTCTGCG CCGCCAACGGTATTGATGCCCCTGATCCAAGAAACACTGGAGGGAATGAACGTCAAATGTAAACTTGACCCTAGA TCTAAGATGGTGCTGCTGTTGAGGATCGGAGGGTACGACAAGCGCAAGCTCAAGTTCCGAGGGTGGGTGAACGTG GAGAGCTTCAGCAAACATGGGTACGAGGGGAGCTTCTGCGGCATGGCGAGAGATGAGGGGAGCCCGATTTCGTGG CGGCAGCTGTGGAAGGCCCTCATCAAGAGTCCCCTGGTCAGCCCCCACGTCCTATTCAAATAGCGCCCGAGCGAG CGCCAGCCATCGAAATCCAGGCTATGTTCATGCTAAATTGGCGGATGGATGAGGATTCTCCCAACTTCGAATTTC TTAATTTTTGAGACGGACGTCGACCCCCCTTATACGTGAGCGCGTTCTTGTTTCTTGTGACCCAACGTTTCGGTA TTTTGTATCATGGAAATCATACCAGGTATCTCCAATGGACTACATGTGTCTCGGACAA |
| Length | 1633 |
Gene
| Sequence id | CopciAB_450167.T0 |
|---|---|
| Sequence |
>CopciAB_450167.T0 AGCGTTTCTTTCTTCTACCACCACAACGGCGCCAAAATGCCCGAATTCCCAAAGGTTACAGGTCAGAATATCGAA TGAAATGTGGTTTAAGGCATTCACTAACAACTGTAATAGGGTACACGCTAGTGGACAGAATTGGAGGCGGCGGGT TCTCCAGGTACGTTGTCCCCGTTCCTTTCAGCTAAAATTCCAATAAAAATCAGCGTTTATAAAGCTATAGACTTC AAGAACTACCGTGCAGCAGCATGTAAAGTGATCATCATTACTGAGGAAACCACAGATAAAGAGAGAAAGACACTC GACAAGGAAATGCGGATCCATTCGGCGCTGAAGCACGAAAACGTGCTGGAATTTCTGAACGCTGTGGTCGTCGAG CTCAAGCACAAACATGCCTATCATCCGGGATATTATATGCTAATGGAGCTGGCTTGTGGTGGAGATCTGTTCGAT AAGATCGGTGGGTAAAGAAGGCTGTGAAATAGACCAGATCTGACAGTTGGTTCCAGCGCCCGACAAGGGCGTCAG TGACGACCTGGCACATCTGTATTTTAACCAGTTGATAGCCGGAATGGTGAGTCTAAACTCCTTTCCGATGAAACC AAGTCTCACAGGTCAAACCAGGACTATATCCACCAACAAGGCGTGTGTCATCGCGACCTTAAGCCGGAGAATCTA CTACTGGACGTCGCTGGGACGCTCAAGATCTCCGATTTTGGTTTATCTTCAGTGTACAAACTGAAAGACTCGGGT CGGACGCGATCTCTGACCGAACGCTGTGGAAGCCTCCCGTATGTTGCACCAGAGGTAGGTGGTCCCGTAAGGTTT GGGCTGTAGACCCAGTGGGCCCACACCCAGCGTTTGATATTCGTTGATCCCGGGGTTGTATCGGGGGTCTCCATT TTCTGGCCCTCGTCCCGTCGGTAGTGTTATTCCTGCGGATGTACCTCATCCCCATATCCCCAGGAATCGTGACGA GTCGAACAGCTGGCAAGGGGGACCCCGTGCTCCACATATCACCGCATATCAAAACTGGGTTGGGGAAAACTGGGG GCATCTGCGATCCGCTCCATTCTGACATATCCCAAAGCTGAACAGCAACGCGCCGTACGAAGCGGAGCCAATCGA TGTCTGGGGATGTGGGGTCATCCTCTTTACTCTGTTGGTCGGTAGTAAGTAATTGGAACGTGTCTTGTGGCACTG GCGCTAAATCGACATCCAGATACCCCATGGGACGAGCCCACCCGACACAGCCCAGAATTCTGCCGCTATCTGGAT GGGACCATCTTCGACGAAGATCCATGGAATCGGATCTCTGAGACCCCGCTGTGTGAGTCCATCAGGCGAACTCCA GCAAGCCCATTCTAAGATGATCTAGCTCTCCTTCAGGGTATGCTGAACGTGAACCCAGCTGAGCGTATGACGCTG TCTGACGTGTATCAGCATCCATGGTGTTTACGGTGAGTTAGTGTTGTGATGCAGTACACTGCTTTTTCTGATCGT TCCGACCAGACCGAGCCAATTGGCACGAGCAGACCCAGGTACCCTGGCGGAGATGCTGACGCAGGGGCTACGCGA GGACGGCGATCTAAATGTAGCAGCCCCTGATTTCGGAGAAGCTATGTCAGTACTTCTCTCGTTTTCCCATCGAGT TTTACTGACCTTTCTTCAGGGATGTCGACGAAGAGGACGATCCCCAGCGTTATGCTGGAACAGCCTCTCAATTCA CGCAGAAACTCATGCTTTTCGTAAGCGTCCCTGCCGCATTGCAGAGAAGGGGAAACTGATCTGTTTCCAGTCGCA AACGCAATCGGGCCCTCGATATGTGCCGAACCTGACTCGATTCTACGCTTCTGCGCCGCCAACGGTATTGATGCC CCTGATCCAAGAAACACTGGAGGGAATGAACGTCAAATGTAAACTTGACCCTAGATCTAAGATGGTGCTGCTGTT GAGGATCGGAGGGTACGACAAGCGCAAGCTCAAGTTCCGAGGGTGGGTGAACGTGGAGAGCTTCAGCAAACATGG GTACGAGGGGAGCTTCTGCGGCATGGCGAGAGATGAGGTTGGCATCACGATAATTCATCGAGGCTTTTCTGACGT TCTTTACAGGGGAGCCCGATTTCGTGGCGGCAGCTGTGGAAGGCCCTCATCAAGAGTCCCCTGGTCAGCCCCCAC GTCCTATTCAAATAGCGCCCGAGCGAGCGCCAGCCATCGAAATCCAGGCTATGTTCATGCTAAATTGGCGGATGG ATGAGGATTCTCCCAACTTCGAATTTCTTAATTTTTGAGACGGACGTCGACCCCCCTTATACGTGAGCGCGTTCT TGTTTCTTGTGACCCAACGTTTCGGTATTTTGTATCATGGAAATCATACCAGGTATCTCCAATGGACTACATGTG TCTCGGACAA |
| Length | 2410 |