CopciAB_450257
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_450257 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 450257 |
| Uniprot id | Functional description | ||
| Location | scaffold_7:54517..56646 | Strand | - |
| Gene length (nt) | 2130 | Transcript length (nt) | 1917 |
| CDS length (nt) | 1821 | Protein length (aa) | 606 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_110545 | 39.1 | 7.385E-103 | 347 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB30858 | 36.4 | 4.386E-83 | 289 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1447359 | 34.6 | 2.176E-72 | 257 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1061463 | 34.1 | 4.043E-71 | 253 |
| Agrocybe aegerita | Agrae_CAA7270120 | 31.4 | 1.08E-68 | 246 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11146 | 29.6 | 1.03E-65 | 237 |
| Lentinula edodes NBRC 111202 | Lenedo1_1165083 | 29.6 | 2.235E-65 | 236 |
| Lentinula edodes B17 | Lened_B_1_1_4422 | 33.4 | 3.807E-65 | 235 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_177485 | 28.3 | 6.454E-55 | 204 |
| Pleurotus ostreatus PC9 | PleosPC9_1_81450 | 36.1 | 3.084E-52 | 196 |
| Grifola frondosa | Grifr_OBZ65583 | 30.2 | 6.544E-50 | 189 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_107374 | 28.5 | 2.342E-48 | 184 |
| Flammulina velutipes | Flave_chr07AA01182 | 25.5 | 1.245E-22 | 103 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_450257.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| No records | |||||
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008266 | Tyrosine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
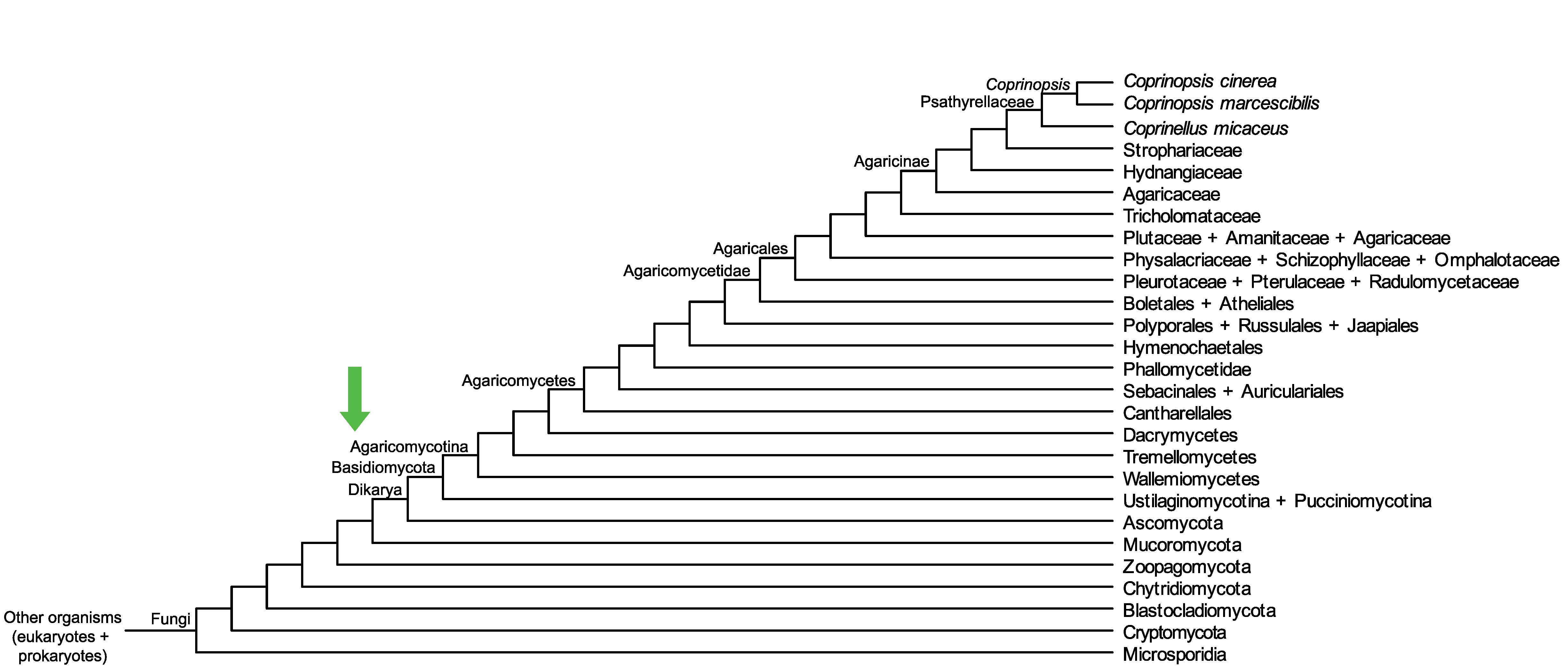
Conservation of CopciAB_450257 across fungi.
Arrow shows the origin of gene family containing CopciAB_450257.
Protein
| Sequence id | CopciAB_450257.T0 |
|---|---|
| Sequence |
>CopciAB_450257.T0 MSVQLNCLLSGLSREDIVQVQVEPGYTIARLRAAICDQLRAQAQARGGRDHLEGTSEIDLQLIGVSLPFDELDKA ARPFEIPGALVWEHYLDRVGASFEGVRDDCVHVVVVRPETVADRDEGRREMSRRRKHEEEEDDFPELTKRLKRTL SVPEKFTPSEGSKPSNYSAIQDGDAPILDGRYRNRTNDLALPVEVYHPVFARFRARAADPALVIPDDMLGLTTEL MRKASRISIGEPESESRTEGMRVLLTEILCFTVVRLGYGNQGWLSPSYVAALPRTEQPLGCAAVVFLEETHEIGQ DGDGTTKGGLAYLRHWSDRNQGGLRERLFCPSFIVSIAGPWIAVLGAVLTTNAIVHRLTDYIWIGESRVLDDANT FRIARVLTALRQSVDDLTTFYESEISMSSNPSRFFPRATSYYDDKRGQKVAFAYLGGLVRYRCAAFMAVVPEERR FIVVKFVERYGVEAHRLLEGKGLAPALLYYGDIWHDDPLAQAGCGTRKMVVMEHVEGRVCVDGVAEGEREVVKEA VRCLHEAGFVHGDIRGQNVLVVGGEGGAKAGSVKLLDFDWAGREGEVRYPLRLSKDLRWPSGVRDNGLILAEYDN EMVDWL |
| Length | 606 |
Coding
| Sequence id | CopciAB_450257.T0 |
|---|---|
| Sequence |
>CopciAB_450257.T0 ATGTCTGTTCAACTTAACTGCCTTCTCTCAGGTCTCAGCCGAGAAGACATCGTCCAGGTCCAGGTCGAACCTGGA TACACCATAGCCCGCCTGCGCGCGGCCATATGCGACCAACTGCGCGCTCAGGCTCAGGCTCGGGGTGGAAGAGAC CACCTCGAGGGGACGTCGGAGATCGACCTCCAGCTTATCGGTGTTTCGCTTCCGTTTGATGAGTTGGACAAGGCG GCAAGGCCGTTTGAGATCCCTGGTGCGTTGGTGTGGGAGCATTATTTGGACCGGGTTGGTGCGTCTTTTGAGGGT GTGCGGGATGATTGTGTGCATGTCGTTGTGGTTCGGCCTGAGACTGTGGCTGACCGTGACGAGGGGAGAAGGGAG ATGTCTCGTAGGCGGAAGCACGAGGAAGAGGAGGACGATTTCCCAGAGTTGACTAAACGTTTAAAAAGAACGCTC TCCGTCCCCGAGAAATTCACACCTTCGGAGGGCAGCAAGCCAAGTAACTACAGCGCGATCCAAGACGGGGATGCG CCGATTTTGGACGGACGATATAGGAATAGGACGAACGATCTCGCCCTCCCTGTGGAGGTCTACCACCCTGTGTTC GCGCGTTTCCGAGCTCGCGCTGCGGACCCCGCACTGGTCATCCCCGACGACATGCTCGGTCTCACGACCGAGCTC ATGCGAAAGGCGTCGCGGATATCGATAGGGGAGCCGGAATCGGAATCCCGCACTGAGGGGATGAGGGTGCTCCTC ACCGAGATCTTGTGCTTCACCGTCGTGCGGTTGGGGTATGGCAACCAGGGCTGGTTGTCACCGAGCTACGTCGCG GCATTACCGCGCACTGAACAACCGCTGGGGTGTGCCGCGGTCGTCTTCCTCGAGGAGACGCATGAGATCGGTCAG GATGGAGATGGGACTACCAAGGGAGGCTTGGCGTACCTGCGTCACTGGAGTGACCGCAATCAAGGGGGTCTCCGG GAAAGGTTGTTCTGTCCGTCGTTCATCGTTTCGATCGCAGGACCGTGGATCGCCGTCCTCGGGGCGGTTTTGACG ACGAACGCCATCGTCCATCGTCTGACGGACTACATCTGGATTGGCGAGAGCCGCGTTCTCGACGACGCCAATACC TTCCGCATCGCCCGTGTGCTCACTGCCTTACGACAATCCGTAGACGATCTGACGACGTTCTACGAATCCGAAATT TCAATGTCCAGCAATCCCTCTCGGTTCTTTCCCCGAGCCACCTCCTACTATGATGATAAGCGCGGACAAAAAGTC GCGTTTGCGTACTTGGGAGGTTTGGTTCGGTATAGGTGTGCGGCGTTTATGGCGGTCGTGCCGGAAGAGAGGCGG TTTATAGTTGTCAAGTTTGTCGAGCGGTACGGTGTGGAGGCGCATAGATTGCTCGAGGGGAAGGGTCTAGCTCCG GCACTGTTGTACTACGGGGATATATGGCACGACGACCCCCTCGCGCAAGCTGGATGCGGTACGAGGAAGATGGTG GTAATGGAGCACGTTGAGGGGAGGGTTTGTGTTGATGGAGTTGCGGAAGGGGAGAGGGAGGTGGTCAAGGAGGCT GTACGGTGTTTGCACGAAGCTGGGTTTGTGCATGGGGATATTCGTGGGCAGAATGTTTTGGTTGTGGGTGGCGAG GGTGGGGCGAAGGCTGGGAGTGTGAAGTTGCTGGACTTTGATTGGGCTGGGAGGGAGGGAGAGGTTAGATATCCT CTTCGCTTGTCGAAGGATCTTCGTTGGCCGAGTGGAGTACGTGATAATGGTCTCATTTTAGCGGAGTATGATAAC GAGATGGTGGATTGGTTGTAG |
| Length | 1821 |
Transcript
| Sequence id | CopciAB_450257.T0 |
|---|---|
| Sequence |
>CopciAB_450257.T0 GTCCTCTGTGCCCCCTTTCTCCTCAATATGTCTGTTCAACTTAACTGCCTTCTCTCAGGTCTCAGCCGAGAAGAC ATCGTCCAGGTCCAGGTCGAACCTGGATACACCATAGCCCGCCTGCGCGCGGCCATATGCGACCAACTGCGCGCT CAGGCTCAGGCTCGGGGTGGAAGAGACCACCTCGAGGGGACGTCGGAGATCGACCTCCAGCTTATCGGTGTTTCG CTTCCGTTTGATGAGTTGGACAAGGCGGCAAGGCCGTTTGAGATCCCTGGTGCGTTGGTGTGGGAGCATTATTTG GACCGGGTTGGTGCGTCTTTTGAGGGTGTGCGGGATGATTGTGTGCATGTCGTTGTGGTTCGGCCTGAGACTGTG GCTGACCGTGACGAGGGGAGAAGGGAGATGTCTCGTAGGCGGAAGCACGAGGAAGAGGAGGACGATTTCCCAGAG TTGACTAAACGTTTAAAAAGAACGCTCTCCGTCCCCGAGAAATTCACACCTTCGGAGGGCAGCAAGCCAAGTAAC TACAGCGCGATCCAAGACGGGGATGCGCCGATTTTGGACGGACGATATAGGAATAGGACGAACGATCTCGCCCTC CCTGTGGAGGTCTACCACCCTGTGTTCGCGCGTTTCCGAGCTCGCGCTGCGGACCCCGCACTGGTCATCCCCGAC GACATGCTCGGTCTCACGACCGAGCTCATGCGAAAGGCGTCGCGGATATCGATAGGGGAGCCGGAATCGGAATCC CGCACTGAGGGGATGAGGGTGCTCCTCACCGAGATCTTGTGCTTCACCGTCGTGCGGTTGGGGTATGGCAACCAG GGCTGGTTGTCACCGAGCTACGTCGCGGCATTACCGCGCACTGAACAACCGCTGGGGTGTGCCGCGGTCGTCTTC CTCGAGGAGACGCATGAGATCGGTCAGGATGGAGATGGGACTACCAAGGGAGGCTTGGCGTACCTGCGTCACTGG AGTGACCGCAATCAAGGGGGTCTCCGGGAAAGGTTGTTCTGTCCGTCGTTCATCGTTTCGATCGCAGGACCGTGG ATCGCCGTCCTCGGGGCGGTTTTGACGACGAACGCCATCGTCCATCGTCTGACGGACTACATCTGGATTGGCGAG AGCCGCGTTCTCGACGACGCCAATACCTTCCGCATCGCCCGTGTGCTCACTGCCTTACGACAATCCGTAGACGAT CTGACGACGTTCTACGAATCCGAAATTTCAATGTCCAGCAATCCCTCTCGGTTCTTTCCCCGAGCCACCTCCTAC TATGATGATAAGCGCGGACAAAAAGTCGCGTTTGCGTACTTGGGAGGTTTGGTTCGGTATAGGTGTGCGGCGTTT ATGGCGGTCGTGCCGGAAGAGAGGCGGTTTATAGTTGTCAAGTTTGTCGAGCGGTACGGTGTGGAGGCGCATAGA TTGCTCGAGGGGAAGGGTCTAGCTCCGGCACTGTTGTACTACGGGGATATATGGCACGACGACCCCCTCGCGCAA GCTGGATGCGGTACGAGGAAGATGGTGGTAATGGAGCACGTTGAGGGGAGGGTTTGTGTTGATGGAGTTGCGGAA GGGGAGAGGGAGGTGGTCAAGGAGGCTGTACGGTGTTTGCACGAAGCTGGGTTTGTGCATGGGGATATTCGTGGG CAGAATGTTTTGGTTGTGGGTGGCGAGGGTGGGGCGAAGGCTGGGAGTGTGAAGTTGCTGGACTTTGATTGGGCT GGGAGGGAGGGAGAGGTTAGATATCCTCTTCGCTTGTCGAAGGATCTTCGTTGGCCGAGTGGAGTACGTGATAAT GGTCTCATTTTAGCGGAGTATGATAACGAGATGGTGGATTGGTTGTAGTATTGGGGTTTCGGCACGTACTCTACA CACAGCTGGTATTTTGGGAATGTAATCTACAAATTCCCTGTC |
| Length | 1917 |
Gene
| Sequence id | CopciAB_450257.T0 |
|---|---|
| Sequence |
>CopciAB_450257.T0 GTCCTCTGTGCCCCCTTTCTCCTCAATATGTCTGTTCAACTTAACTGCCTTCTCTCAGGTCTCAGCCGAGAAGAC ATCGTCCAGGTCCAGGTCGAACCTGGATACACCATAGCCCGCCTGCGCGCGGCCATATGCGACCAACTGCGCGCT CAGGCTCAGGCTCGGGGTGGAAGAGACCACCTCGAGGGGACGTCGGAGATCGACCTCCAGCTTATCGGTGTTTCG CTTCCGTTTGATGAGTTGGACAAGGCGGCAAGGCCGTTTGAGATCCCTGGTGCGTTGGTGTGGGAGCATTATTTG GACCGGGTTGGTGCGTCTTTTGAGGGTGTGCGGGATGATTGTGTGCATGTCGTTGTGGTTCGGCCTGAGACTGTG GCTGACCGTGACGGTGAGCGGTTTTTTTATATAACTACTAACTAGGATCTGAGTCGTTTTGGGCGTCTGAACTTA TTCTTTTTTCGCAGAGGGGAGAAGGGAGATGTCTCGTAGGCGGAAGCACGAGGAAGAGGAGGACGATTTCCCAGA GTTGACTAAACGTGCGTTTATCCAATCCTCCATCTCATCCCAACCAAGCCTCATCACCTCGGTCTCTTCAGGTTT AAAAAGAACGCTCTCCGTCCCCGAGAAATTCACACCTTCGGAGGGCAGCAAGCCAAGTAACTACAGCGCGATCCA AGACGGGGATGCGCCGATTTTGGACGGACGATATAGGAATAGGACGAACGATCTCGCCCTCCCTGTGGAGGTCTA CCACCCTGTGTTCGCGCGTTTCCGAGCTCGCGCTGCGGACCCCGCACTGGTCATCCCCGACGACATGCTCGGTCT CACGACCGAGCTCATGCGAAAGGCGTCGCGGATATCGATAGGGGAGCCGGAATCGGAATCCCGCACTGAGGGGAT GAGGGTGCTCCTCACCGAGATCTTGTGCTTCACCGTCGTGCGGTTGGGGTATGGCAACCAGGGCTGGTTGTCACC GAGCTACGTCGCGGCATTACCGCGCACTGAACAACCGCTGGGGTGTGCCGCGGTCGTCTTCCTCGAGGAGACGCA TGAGATCGGTCAGGATGGAGATGGGACTACCAAGGGAGGCTTGGCGTACCTGCGTCACTGGAGTGACCGCAATCA AGGGGTAAGTGTTCCATGCTCTCTATCATAGATGATGAGAATTTGACCAAGTTAAATACATCGCGTTTTGCTAAC TCCTAGGGTCTCCGGGAAAGGTTGTTCTGTCCGTCGTTCATCGTTTCGATCGCAGGACCGTGGATCGCCGTCCTC GGGGCGGTTTTGACGACGAACGCCATCGTCCATCGTCTGACGGACTACATCTGGATTGGCGAGAGCCGCGTTCTC GACGACGCCAATACCTTCCGCATCGCCCGTGTGCTCACTGCCTTACGACAATCCGTAGACGATCTGACGACGTTC TACGAATCCGAAATTTCAATGTCCAGCAATCCCTCTCGGTTCTTTCCCCGAGCCACCTCCTACTATGATGATAAG CGCGGACAAAAAGTCGCGTTTGCGTACTTGGGAGGTTTGGTTCGGTATAGGTGTGCGGCGTTTATGGCGGTCGTG CCGGAAGAGAGGCGGTTTATAGTTGTCAAGTTTGTCGAGCGGTACGGTGTGGAGGCGCATAGATTGCTCGAGGGG AAGGGTCTAGCTCCGGCACTGTTGTACTACGGGGATATATGGCACGACGACCCCCTCGCGCAAGCTGGATGCGGT ACGAGGAAGATGGTGGTAATGGAGCACGTTGAGGGGAGGGTTTGTGTTGATGGAGTTGCGGAAGGGGAGAGGGAG GTGGTCAAGGAGGCTGTACGGTGTTTGCACGAAGCTGGGTTTGTGCATGGGGATATTCGTGGGCAGAATGTTTTG GTTGTGGGTGGCGAGGGTGGGGCGAAGGCTGGGAGTGTGAAGTTGCTGGACTTTGATTGGGCTGGGAGGGAGGGA GAGGTTAGATATCCTCTTCGCTTGTCGAAGGATCTTCGTTGGCCGAGTGGAGTACGTGATAATGGTCTCATTTTA GCGGAGTATGATAACGAGATGGTGGATTGGTTGTAGTATTGGGGTTTCGGCACGTACTCTACACACAGCTGGTAT TTTGGGAATGTAATCTACAAATTCCCTGTC |
| Length | 2130 |