CopciAB_451588
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_451588 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 451588 |
| Uniprot id | Functional description | Belongs to the GMC oxidoreductase family | |
| Location | scaffold_11:387866..390673 | Strand | - |
| Gene length (nt) | 2808 | Transcript length (nt) | 2125 |
| CDS length (nt) | 1830 | Protein length (aa) | 609 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7263638 | 72.3 | 3.833E-299 | 917 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29937 | 69.5 | 7.995E-292 | 896 |
| Grifola frondosa | Grifr_OBZ79543 | 63.4 | 4.863E-249 | 772 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_112928 | 61.8 | 1.079E-248 | 771 |
| Flammulina velutipes | Flave_chr01AA00453 | 60.3 | 7.504E-244 | 757 |
| Lentinula edodes B17 | Lened_B_1_1_7642 | 59.2 | 6.432E-244 | 757 |
| Lentinula edodes NBRC 111202 | Lenedo1_1059754 | 59.2 | 1.802E-243 | 756 |
| Auricularia subglabra | Aurde3_1_1337256 | 60.4 | 7.042E-242 | 752 |
| Schizophyllum commune H4-8 | Schco3_2641528 | 59.5 | 4.387E-234 | 729 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_18550 | 59.1 | 9.553E-184 | 583 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_451588.T0 |
| Description | Belongs to the GMC oxidoreductase family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 41 | 354 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 457 | 600 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 20 | 0.9764 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
| IPR027424 | Glucose Oxidase, domain 2 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| E | Belongs to the GMC oxidoreductase family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
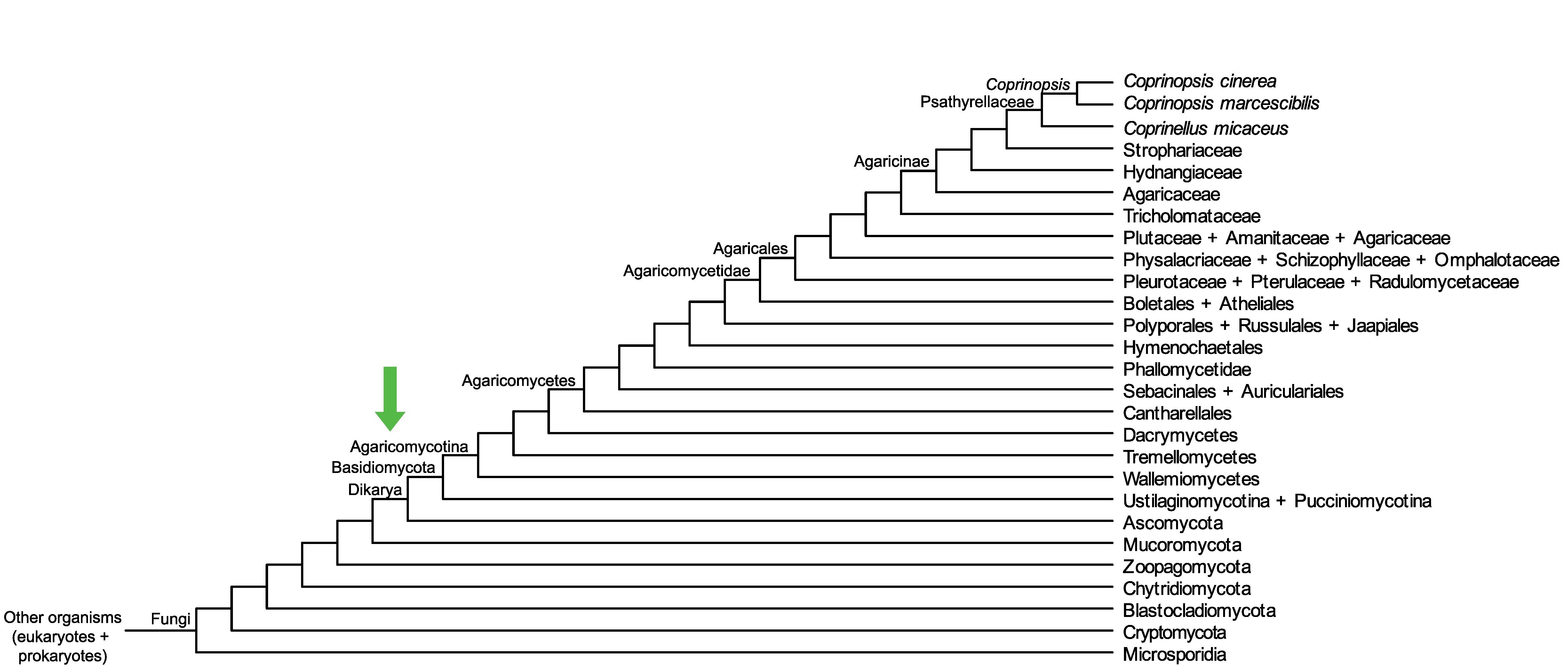
Conservation of CopciAB_451588 across fungi.
Arrow shows the origin of gene family containing CopciAB_451588.
Protein
| Sequence id | CopciAB_451588.T0 |
|---|---|
| Sequence |
>CopciAB_451588.T0 MPSFSAVTLLALSLVSVASGASPLQKRGVTTSPDAVNGQSYDYIVVGGGLAGITVAARLAENPSVTVLVIEAGGD DRNDPRVYDIYRYGEAFGSELMWDWPTERNRGIVGGKTLGGGTSINGAAFTRGVDAQYDAWSALLEPWEANVGWN WQGMFEYMKKSEAFSPPNSEQMSKGAQFVDWYHGYGGPVQVTNPNSMYGGPHQWFFIDSMVGLTGMNHIPDVNGG NPNGVSITPLMINWRDADHRSSSVMAYLTPVEHQRTNWVTLTRHFVTKINWSNNSVPFRASGVEFGPANGGSTRY TAFARREVILAAGAIQTPALLQLSGIGDSAILGPLGIQTHVDLKTVGKNLQEQTMSQFGAGGNGFNPEGRGPSNV IAFPNLYQLFGTQASSIVNRIRSSISSWAASQAGSGFSAAALQQIFQVQADLIINNNAPVAELFFSYGWPDTLGI QMWNLLPFSRGSVQITSNNPYTKPRVQVNFFDIDLDMDIQVAASRLCRRALTTPPLSWIATGETIPGNAVPDNAE RGTDNDWKNWINNNFNTVSHPVGTAAMMRRSLGGVVDAQLRVYDTANVRVVDASVLALQVSAHLSSTLYGIAEKA ADLIKATYP |
| Length | 609 |
Coding
| Sequence id | CopciAB_451588.T0 |
|---|---|
| Sequence |
>CopciAB_451588.T0 ATGCCCTCCTTCAGCGCTGTCACTCTTTTGGCGCTCTCCCTTGTTTCCGTCGCGTCTGGCGCGTCTCCCCTGCAA AAGCGCGGCGTGACGACTTCACCCGACGCCGTGAATGGTCAGTCCTACGACTATATTGTCGTCGGTGGTGGCTTA GCAGGGATTACCGTTGCTGCTCGATTGGCTGAGAACCCATCGGTCACCGTTCTGGTCATCGAAGCAGGGGGTGAT GACAGGAATGATCCCCGGGTGTACGATATTTATCGTTACGGGGAAGCCTTTGGCTCGGAGCTGATGTGGGATTGG CCAACCGAGCGAAATCGAGGCATCGTCGGTGGTAAAACGCTCGGAGGAGGGACTTCGATCAATGGTGCTGCCTTC ACTCGAGGAGTAGACGCCCAATATGACGCGTGGTCGGCCCTTCTCGAACCTTGGGAAGCCAACGTTGGTTGGAAT TGGCAAGGCATGTTTGAGTACATGAAGAAGTCCGAAGCCTTTTCGCCTCCCAACAGCGAGCAAATGTCGAAGGGC GCTCAGTTTGTTGACTGGTACCATGGCTACGGGGGCCCCGTTCAAGTGACCAACCCTAATAGCATGTATGGCGGC CCTCATCAATGGTTCTTTATTGACTCCATGGTCGGCTTGACTGGCATGAATCACATCCCAGATGTGAATGGAGGC AACCCCAATGGCGTCTCGATTACTCCTCTGATGATTAATTGGCGCGATGCTGACCACCGATCTTCGTCCGTTATG GCCTATCTGACGCCAGTTGAGCATCAGCGTACAAACTGGGTTACTTTGACGAGGCACTTTGTCACTAAAATCAAT TGGTCAAATAACTCTGTGCCTTTCAGAGCCTCTGGTGTCGAATTTGGTCCTGCCAATGGTGGCTCGACGAGGTAT ACTGCTTTTGCCAGACGCGAAGTCATCTTGGCTGCCGGCGCCATCCAGACCCCTGCCCTCCTTCAACTTTCAGGA ATCGGCGACTCTGCCATCCTCGGGCCTCTCGGAATCCAAACCCACGTCGACCTCAAGACAGTTGGCAAGAACCTT CAGGAACAGACCATGAGTCAATTTGGTGCAGGTGGCAATGGCTTCAATCCTGAAGGTCGTGGCCCTTCGAACGTC ATCGCGTTCCCGAATCTGTACCAGCTGTTCGGTACCCAAGCAAGCTCAATCGTCAACCGTATTAGGTCGTCGATT AGCTCTTGGGCTGCGTCACAGGCTGGCTCAGGGTTCTCAGCTGCTGCTCTCCAACAGATTTTCCAGGTGCAGGCA GATTTGATCATCAACAATAACGCACCGGTTGCAGAGCTGTTCTTCAGCTATGGATGGCCCGATACTCTGGGTATT CAGATGTGGAATCTCTTGCCATTCAGTCGAGGCAGCGTTCAGATCACCAGCAACAACCCCTACACCAAGCCACGC GTTCAAGTCAATTTCTTCGATATCGACTTGGACATGGATATCCAAGTTGCGGCGTCACGTCTTTGCCGACGTGCT CTGACCACCCCGCCCTTGAGTTGGATCGCCACTGGAGAAACCATCCCAGGCAACGCCGTCCCAGATAACGCAGAA CGAGGAACAGACAACGACTGGAAGAACTGGATCAACAACAACTTCAACACTGTCTCCCATCCCGTCGGAACGGCG GCTATGATGAGACGCAGCCTTGGAGGTGTCGTGGATGCCCAACTCAGGGTGTATGACACCGCCAACGTCCGCGTT GTGGATGCTTCTGTGCTCGCCCTCCAGGTTAGCGCCCACCTGTCATCCACTCTCTACGGTATTGCGGAGAAGGCG GCGGACCTCATCAAGGCTACGTACCCTTGA |
| Length | 1830 |
Transcript
| Sequence id | CopciAB_451588.T0 |
|---|---|
| Sequence |
>CopciAB_451588.T0 ACCCGGTTCTTTTCCCTCTCTTTTTTCTCCTTTTTTTTCGCCATGCCCTCCTTCAGCGCTGTCACTCTTTTGGCG CTCTCCCTTGTTTCCGTCGCGTCTGGCGCGTCTCCCCTGCAAAAGCGCGGCGTGACGACTTCACCCGACGCCGTG AATGGTCAGTCCTACGACTATATTGTCGTCGGTGGTGGCTTAGCAGGGATTACCGTTGCTGCTCGATTGGCTGAG AACCCATCGGTCACCGTTCTGGTCATCGAAGCAGGGGGTGATGACAGGAATGATCCCCGGGTGTACGATATTTAT CGTTACGGGGAAGCCTTTGGCTCGGAGCTGATGTGGGATTGGCCAACCGAGCGAAATCGAGGCATCGTCGGTGGT AAAACGCTCGGAGGAGGGACTTCGATCAATGGTGCTGCCTTCACTCGAGGAGTAGACGCCCAATATGACGCGTGG TCGGCCCTTCTCGAACCTTGGGAAGCCAACGTTGGTTGGAATTGGCAAGGCATGTTTGAGTACATGAAGAAGTCC GAAGCCTTTTCGCCTCCCAACAGCGAGCAAATGTCGAAGGGCGCTCAGTTTGTTGACTGGTACCATGGCTACGGG GGCCCCGTTCAAGTGACCAACCCTAATAGCATGTATGGCGGCCCTCATCAATGGTTCTTTATTGACTCCATGGTC GGCTTGACTGGCATGAATCACATCCCAGATGTGAATGGAGGCAACCCCAATGGCGTCTCGATTACTCCTCTGATG ATTAATTGGCGCGATGCTGACCACCGATCTTCGTCCGTTATGGCCTATCTGACGCCAGTTGAGCATCAGCGTACA AACTGGGTTACTTTGACGAGGCACTTTGTCACTAAAATCAATTGGTCAAATAACTCTGTGCCTTTCAGAGCCTCT GGTGTCGAATTTGGTCCTGCCAATGGTGGCTCGACGAGGTATACTGCTTTTGCCAGACGCGAAGTCATCTTGGCT GCCGGCGCCATCCAGACCCCTGCCCTCCTTCAACTTTCAGGAATCGGCGACTCTGCCATCCTCGGGCCTCTCGGA ATCCAAACCCACGTCGACCTCAAGACAGTTGGCAAGAACCTTCAGGAACAGACCATGAGTCAATTTGGTGCAGGT GGCAATGGCTTCAATCCTGAAGGTCGTGGCCCTTCGAACGTCATCGCGTTCCCGAATCTGTACCAGCTGTTCGGT ACCCAAGCAAGCTCAATCGTCAACCGTATTAGGTCGTCGATTAGCTCTTGGGCTGCGTCACAGGCTGGCTCAGGG TTCTCAGCTGCTGCTCTCCAACAGATTTTCCAGGTGCAGGCAGATTTGATCATCAACAATAACGCACCGGTTGCA GAGCTGTTCTTCAGCTATGGATGGCCCGATACTCTGGGTATTCAGATGTGGAATCTCTTGCCATTCAGTCGAGGC AGCGTTCAGATCACCAGCAACAACCCCTACACCAAGCCACGCGTTCAAGTCAATTTCTTCGATATCGACTTGGAC ATGGATATCCAAGTTGCGGCGTCACGTCTTTGCCGACGTGCTCTGACCACCCCGCCCTTGAGTTGGATCGCCACT GGAGAAACCATCCCAGGCAACGCCGTCCCAGATAACGCAGAACGAGGAACAGACAACGACTGGAAGAACTGGATC AACAACAACTTCAACACTGTCTCCCATCCCGTCGGAACGGCGGCTATGATGAGACGCAGCCTTGGAGGTGTCGTG GATGCCCAACTCAGGGTGTATGACACCGCCAACGTCCGCGTTGTGGATGCTTCTGTGCTCGCCCTCCAGGTTAGC GCCCACCTGTCATCCACTCTCTACGGTATTGCGGAGAAGGCGGCGGACCTCATCAAGGCTACGTACCCTTGAAGA AGCGGAAGGGCAACGTTCTGTGTTCCATCCAGTGCTCCTTGTCGGACACATTCTACTTTATACCTATCTATCGGA CATTTTGAAGCCACGGACCTATGTATCCACCCCACACAACCGAAACGTTCTTTCCCAACTTCCCTAACCACTCGT TTTATCTTCACGGACACACTGTACACGTAGAAAACTTTGTAATTCTGGATTGTATAAATCGTGCTAACATAGTAC AATTTGAAGGACAAACGTTCATTTC |
| Length | 2125 |
Gene
| Sequence id | CopciAB_451588.T0 |
|---|---|
| Sequence |
>CopciAB_451588.T0 ACCCGGTTCTTTTCCCTCTCTTTTTTCTCCTTTTTTTTCGCCATGCCCTCCTTCAGCGCTGTCACTCTTTTGGCG CTCTCCCTTGTTTCCGTCGCGTCTGGCGCGTCTCCCCTGCAAAAGCGCGGCGTGACGACTTCACCCGACGCCGTG AATGGTCAGTCCTACGACTATATTGTCGTCGGTGGTGGCTTAGCAGGGATTACCGTTGCTGCTCGATTGGCTGAG AACCCATCGGTCACCGTTCTGGTCATCGAAGCAGGGGGTGATGACAGGAATGATCCCCGGGTGTACGATATTTAT CGTTACGGGGAAGCCTTTGGCTCGGAGCTGATGTGGGATTGGCCAACCGAGCGAAATCGAGGCATCGTCGGTGGT AAAACGCTCGGAGGAGGGAGTACGTACCTCCCTTACCTTATTATTGAGGGAGAAGACTGAACCAGGGACAATAAT AGCTTCGATCAATGGTGCTGCCTTCACTCGAGGAGTAGACGCCCAATATGACGCGTGGTCGGCCCTTCTCGAACC TTGGGAAGCCAACGTTGGTTGGAATTGGCAAGGCATGTTTGAGTACATGAAGAAGGTTCGCTCTTTCCCGTATAA TTACAACACGGCGTTCCTGATTCAATTCACCTCTGCAGTCCGAAGCCTTTTCGCCTCCCAACAGCGAGCAAATGT CGAAGGGCGCTCAGTTTGTTGACTGGTACCATGGCTACGGGGGCCCCGTTCAAGTGACCAACCCTAATAGCATGT ATGGCGGCCCTCATCAATGGTTCTTTATTGACTCCATGGTCGGCTTGACTGGCATGAATCACATCCCAGATGTGA ATGGAGGCAACCCCAATGGCGTCTCGATTACTCCTCTGGTAAGCCTTCCGCAATCACCACAGACCTTGTTGCAAG TCTTACTGAACATGCCCTTCAGATGATTAATTGGCGCGATGCTGACCACCGATCTTCGTCCGTTATGGCCTATCT GACGCCAGTTGAGCATCAGCGTACAAACTGGGTTACTTTGACGAGGCACTTTGTCAGTTTTACCTCACTCGGTCT CTTGGAGGTGGCTGACGCCCCTTTGCTATAGGTCACTAAAATCAATTGGTCAAATAACTCTGTGCCTTTCAGAGC CTCTGGTGTCGAATTTGGTCCTGCCAATGGTGGCTCGACGAGGTATACTGCTTTTGCCAGACGCGAAGTCATCTT GGCTGCCGGCGCCATCCAGGTAAGACGTTCGAGAATCAAAAGTCTATTCGCACACACTTAAATCGTCTACCTCTC AGACCCCTGCCCTCCTTCAACTTTCAGGAATCGGCGACTCTGCCATCCTCGGGCCTCTCGGAATCCAAACCCACG TCGACCTCAAGACAGTTGGCAAGAACCTTCAGGAACAGGTACACCAACTCCACCCTTCCCCTTCGATCACCTCTC TCTAACTGACTGCTGTTTTACCACTTTCTACTCTTTACGCAACCATCTATGTACTACTAAGACCATGAGTCAATT TGGTGCAGGTGGCAATGGCTTCAATCCTGAAGGTCGTGGCCCTTCGAACGTCATCGCGTTCCCGAATCTGTACCA GCTGTTCGGTACCCAAGCAAGCTCAATCGTCAACCGTATTAGGTCGTCGATTAGCTCTTGGGCTGCGTCACAGGC TGGCTCAGGGTTCTCAGCTGCTGCTCTCCAACAGATTTTCCAGGTGCAGGCAGATTTGATCATCAACAATAACGG TACGTCTTCATCTTCACCGACGACGCGTGGCGAACATTGTGCTGACTGTCCTTTGTAGCACCGGTTGCAGAGCTG TTCTTCAGCTATGGATGGCCCGAGTACGTATTCCTTCAACACTTGAAAGATTCTCATTTGTGCTGATTGTTGCCA TAGTACTCTGGGTATTCAGATGTGGAATCTCTTGCCATTCAGTCGAGGCAGCGTTCAGATCACCGTAAGCCTCCT CTCTGTATGTACGTCAGCGGAGTACCTAAAAGCAGTCCGCACCGATAGAGCAACAACCCCTACACCAAGCCACGC GTTCAAGTCAATTTCTTCGATATCGACTTGGACATGGATATCCAAGTTGCGGCGTCACGTCTTTGCCGACGTGCT CTGACCACCCCGCCCTTGAGGTAACCAACTCTTTCCCCTTTTACCACTGGTTGTCCAGGTCGCTGATACCAGTGT TAAGTTGGATCGCCACTGGAGAAACCATCCCAGGCAACGCCGTCCCAGATAACGCAGAACGAGGAACAGACAACG ACTGGAAGAACTGGATCAACAACAACTTCAACACTGTCTCCCATCCCGTCGGAACGGCGGCTATGATGAGACGCA GCCTTGGAGGTACCACTCTCTCCTAGCTTCAAAGCGAAATAAAGACGTAAACTAACCTAAAACCTAATCGCGCAG GTGTCGTGGATGCCCAACTCAGGGTGTATGACACCGCCAACGTCCGCGTTGTGGATGCTTCTGTGCTCGCCCTCC AGGTTAGCGCCCACCTGTCATCCACTCTCTACGGTATTGCGGAGAAGGCGGCGGACCTCATCAAGGCTACGTACC CTTGAAGAAGCGGAAGGGCAACGTTCTGTGTTCCATCCAGTGCTCCTTGTCGGACACATTCTACTTTATACCTAT CTATCGGACATTTTGAAGCCACGGACCTATGTATCCACCCCACACAACCGAAACGTTCTTTCCCAACTTCCCTAA CCACTCGTTTTATCTTCACGGACACACTGTACACGTAGAAAACTTTGTAATTCTGGATTGTATAAATCGTGCTAA CATAGTACAATTTGAAGGACAAACGTTCATTTC |
| Length | 2808 |