CopciAB_451611
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_451611 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | srk1 | Synonyms | 451611 |
| Uniprot id | Functional description | Serine/Threonine protein kinases, catalytic domain | |
| Location | scaffold_5:1304535..1307324 | Strand | + |
| Gene length (nt) | 2790 | Transcript length (nt) | 2228 |
| CDS length (nt) | 1872 | Protein length (aa) | 623 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YLR248W_RCK2 |
| Neurospora crassa | camk-4 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7259533 | 72 | 7.075E-290 | 891 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB22618 | 68.9 | 1.096E-267 | 827 |
| Schizophyllum commune H4-8 | Schco3_2623509 | 67 | 4.666E-259 | 802 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1385570 | 65.7 | 2.218E-257 | 797 |
| Lentinula edodes NBRC 111202 | Lenedo1_1082358 | 67.9 | 1.179E-255 | 792 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_106452 | 65.8 | 4.465E-255 | 790 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191944 | 65.8 | 2.293E-253 | 785 |
| Flammulina velutipes | Flave_chr02AA00105 | 74.5 | 2.152E-247 | 768 |
| Auricularia subglabra | Aurde3_1_1305163 | 62.8 | 4.109E-246 | 765 |
| Pleurotus ostreatus PC9 | PleosPC9_1_83587 | 66 | 2.522E-244 | 759 |
| Pleurotus ostreatus PC15 | PleosPC15_2_61429 | 65.8 | 2.804E-243 | 756 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_23139 | 65.5 | 4.185E-240 | 747 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_114705 | 65.6 | 4.984E-237 | 738 |
| Grifola frondosa | Grifr_OBZ71808 | 68.2 | 1.358E-224 | 702 |
| Lentinula edodes B17 | Lened_B_1_1_12846 | 72 | 9.839E-120 | 397 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | srk1 |
|---|---|
| Protein id | CopciAB_451611.T0 |
| Description | Serine/Threonine protein kinases, catalytic domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14096 | STKc_RCK1-like | - | 95 | 400 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 97 | 400 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR030616 | Aurora kinase-like |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| K08286 |
EggNOG
| COG category | Description |
|---|---|
| T | Serine/Threonine protein kinases, catalytic domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
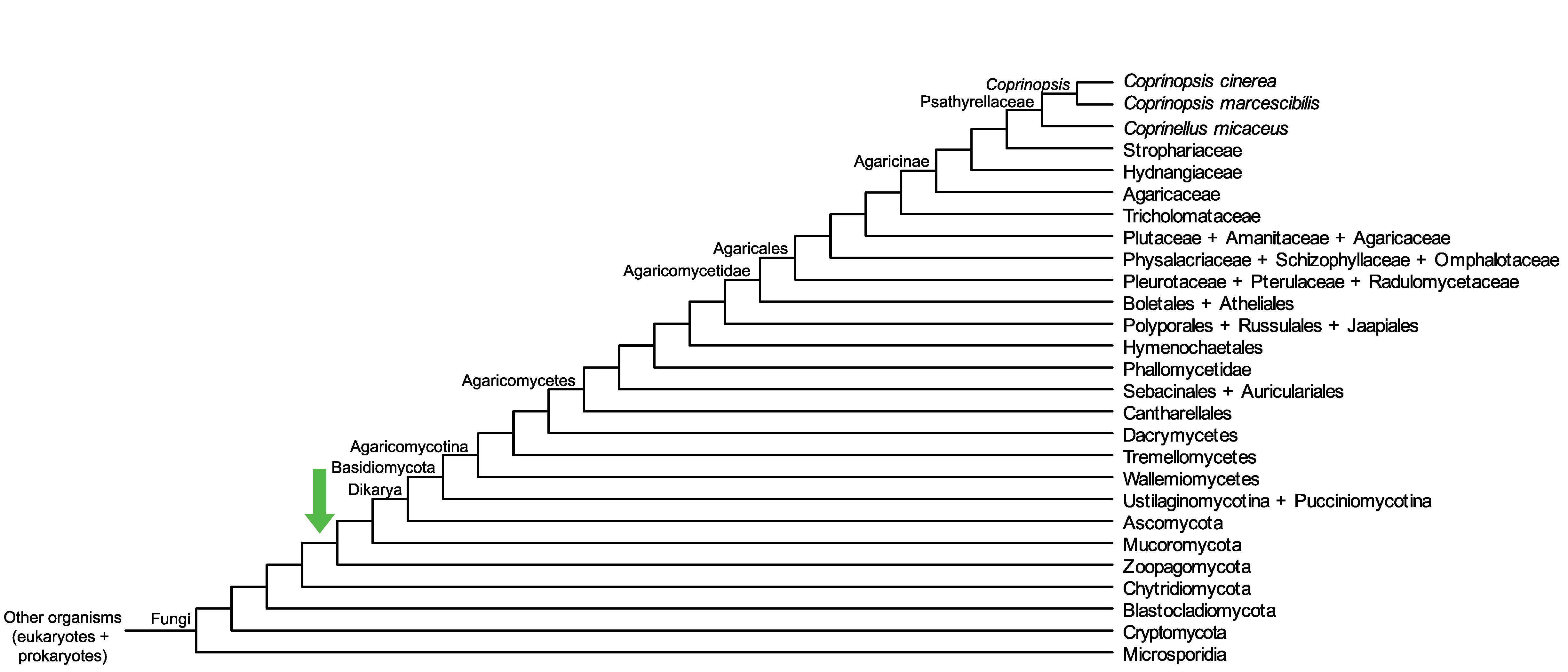
Conservation of CopciAB_451611 across fungi.
Arrow shows the origin of gene family containing CopciAB_451611.
Protein
| Sequence id | CopciAB_451611.T0 |
|---|---|
| Sequence |
>CopciAB_451611.T0 MPPTGVVDAFKNLIRHGKHHNHVQPPRQNEQPRQHQPQGHNGPAPAQASKRHNEQPTAQAAQPHEKSPPPREAAE MIVNQERAEKSKMPTYKGLESYTLLEKMGDGAFSNVYKAVEHSTNRKVAVKVVRKYELNASQAGEKHLNPQFKKK PRVTERANILKEVQIMRGIQHPSIVKLYSFSESPEHYFLVLELMEGGELFHQIVKLTYFSENLSRHVILQVAHGI RHLHEERGVVHRDIKPENLLFERIDIIPSKNPIKRPYDEEKEDEGEFIPGVGGGGIGRVKIADFGLSKVVWDEQT MTPCGTVGYTAPEIVKDERYSKSVDMWALGCVLYTLLCGFPPFYDESINVLTEKVARGYYTFLSPWWDDISESAK DLIKHLLCVDPAQRYTIDEFLAHPWCNAAPAPPPPPTPLVYSDKFSISAAGGRPIDSPLLSAPRGNWEGRSPGLA TLKEAFDITYAVHRMEEEGARRRKYGRGTRGFLSELNEDDEMDDDEDDDITDENDVRRHPGQQQQQPRHPQGQAP QNTSLMEGRAGPRDQGNREWHHAQSQGVGRTSGPSASVARTGANRARGHKGRQFELDLDGATLLERRQRRAAAKK DYPGSPLRMEGLRVDDRPSVTAL |
| Length | 623 |
Coding
| Sequence id | CopciAB_451611.T0 |
|---|---|
| Sequence |
>CopciAB_451611.T0 ATGCCTCCAACCGGCGTCGTCGATGCTTTCAAGAACCTCATTCGCCACGGCAAGCACCACAACCACGTACAGCCG CCCAGACAGAACGAACAGCCTCGTCAGCATCAGCCTCAAGGCCACAACGGCCCCGCCCCTGCCCAGGCGAGCAAA CGCCACAATGAGCAACCCACAGCACAGGCTGCTCAGCCCCACGAAAAGTCGCCACCCCCACGAGAAGCAGCTGAA ATGATCGTCAACCAGGAACGCGCTGAAAAGTCAAAGATGCCAACTTACAAAGGCCTTGAGAGCTATACTTTGCTC GAGAAGATGGGAGATGGAGCATTCTCTAATGTCTACAAGGCTGTGGAGCACTCGACCAACCGCAAAGTTGCTGTC AAGGTCGTCCGTAAATACGAACTCAATGCGTCACAGGCAGGGGAGAAACATCTCAATCCCCAATTCAAGAAGAAG CCGCGTGTGACAGAGCGGGCGAATATTCTCAAAGAGGTGCAGATCATGCGGGGCATTCAGCATCCCTCGATCGTG AAGCTTTACTCCTTTTCAGAATCGCCGGAGCATTACTTCCTTGTGCTTGAGTTAATGGAGGGTGGAGAACTATTC CATCAAATCGTCAAGCTCACCTACTTCAGTGAAAACCTCTCGCGTCATGTTATCCTCCAAGTCGCTCATGGCATA CGACATCTCCATGAAGAACGGGGTGTCGTCCATCGCGATATCAAACCTGAAAACTTGCTATTCGAGCGCATAGAC ATCATCCCTTCCAAGAACCCAATAAAGCGACCGTACGATGAAGAGAAGGAAGACGAAGGCGAGTTCATACCCGGC GTTGGAGGAGGCGGTATTGGTCGCGTCAAGATCGCCGACTTTGGTCTTAGTAAGGTGGTCTGGGATGAGCAGACG ATGACACCCTGTGGTACCGTTGGTTACACCGCACCGGAGATCGTCAAGGATGAGCGATACAGCAAGAGTGTAGAT ATGTGGGCTCTTGGCTGTGTCCTCTACACCCTTCTTTGTGGATTCCCTCCATTCTACGATGAGAGTATCAATGTT CTCACTGAGAAGGTGGCACGGGGATACTATACCTTCCTCAGCCCTTGGTGGGACGATATCAGCGAGAGCGCAAAG GACCTCATCAAGCATCTTCTTTGCGTCGATCCAGCCCAGAGGTATACTATCGACGAATTCCTCGCACACCCTTGG TGCAACGCGGCGCCTGCTCCTCCGCCTCCCCCCACCCCTCTTGTTTACAGCGACAAATTCTCCATATCGGCGGCT GGCGGCCGACCTATTGATTCACCCCTTCTTTCCGCACCCCGGGGTAACTGGGAGGGTCGAAGTCCGGGTCTAGCC ACCTTGAAGGAGGCCTTTGACATTACCTACGCTGTGCATCGCATGGAAGAGGAAGGAGCCAGGCGACGCAAGTAC GGTAGAGGGACCCGGGGCTTCCTTAGCGAGCTTAACGAAGATGACGAGATGGACGATGATGAGGACGATGACATC ACCGATGAAAACGATGTTCGCAGGCACCCAGGACAGCAGCAGCAGCAACCACGTCACCCTCAAGGCCAAGCTCCG CAAAATACGTCGCTAATGGAAGGCCGCGCAGGACCAAGGGATCAAGGGAACCGAGAGTGGCATCACGCACAGTCG CAGGGAGTTGGGAGGACGAGCGGGCCGAGCGCTTCGGTTGCTAGAACGGGTGCGAACAGGGCGAGGGGACACAAA GGTCGACAGTTTGAACTGGATCTGGACGGTGCGACTCTATTGGAGAGGAGGCAGCGACGGGCAGCAGCAAAGAAG GATTATCCGGGTAGCCCATTGCGGATGGAAGGACTCAGGGTGGACGATCGTCCATCTGTGACGGCCCTTTAG |
| Length | 1872 |
Transcript
| Sequence id | CopciAB_451611.T0 |
|---|---|
| Sequence |
>CopciAB_451611.T0 AATTCATCCTTTTTCGATCCCCCATCTCCACAATGCCTCCAACCGGCGTCGTCGATGCTTTCAAGAACCTCATTC GCCACGGCAAGCACCACAACCACGTACAGCCGCCCAGACAGAACGAACAGCCTCGTCAGCATCAGCCTCAAGGCC ACAACGGCCCCGCCCCTGCCCAGGCGAGCAAACGCCACAATGAGCAACCCACAGCACAGGCTGCTCAGCCCCACG AAAAGTCGCCACCCCCACGAGAAGCAGCTGAAATGATCGTCAACCAGGAACGCGCTGAAAAGTCAAAGATGCCAA CTTACAAAGGCCTTGAGAGCTATACTTTGCTCGAGAAGATGGGAGATGGAGCATTCTCTAATGTCTACAAGGCTG TGGAGCACTCGACCAACCGCAAAGTTGCTGTCAAGGTCGTCCGTAAATACGAACTCAATGCGTCACAGGCAGGGG AGAAACATCTCAATCCCCAATTCAAGAAGAAGCCGCGTGTGACAGAGCGGGCGAATATTCTCAAAGAGGTGCAGA TCATGCGGGGCATTCAGCATCCCTCGATCGTGAAGCTTTACTCCTTTTCAGAATCGCCGGAGCATTACTTCCTTG TGCTTGAGTTAATGGAGGGTGGAGAACTATTCCATCAAATCGTCAAGCTCACCTACTTCAGTGAAAACCTCTCGC GTCATGTTATCCTCCAAGTCGCTCATGGCATACGACATCTCCATGAAGAACGGGGTGTCGTCCATCGCGATATCA AACCTGAAAACTTGCTATTCGAGCGCATAGACATCATCCCTTCCAAGAACCCAATAAAGCGACCGTACGATGAAG AGAAGGAAGACGAAGGCGAGTTCATACCCGGCGTTGGAGGAGGCGGTATTGGTCGCGTCAAGATCGCCGACTTTG GTCTTAGTAAGGTGGTCTGGGATGAGCAGACGATGACACCCTGTGGTACCGTTGGTTACACCGCACCGGAGATCG TCAAGGATGAGCGATACAGCAAGAGTGTAGATATGTGGGCTCTTGGCTGTGTCCTCTACACCCTTCTTTGTGGAT TCCCTCCATTCTACGATGAGAGTATCAATGTTCTCACTGAGAAGGTGGCACGGGGATACTATACCTTCCTCAGCC CTTGGTGGGACGATATCAGCGAGAGCGCAAAGGACCTCATCAAGCATCTTCTTTGCGTCGATCCAGCCCAGAGGT ATACTATCGACGAATTCCTCGCACACCCTTGGTGCAACGCGGCGCCTGCTCCTCCGCCTCCCCCCACCCCTCTTG TTTACAGCGACAAATTCTCCATATCGGCGGCTGGCGGCCGACCTATTGATTCACCCCTTCTTTCCGCACCCCGGG GTAACTGGGAGGGTCGAAGTCCGGGTCTAGCCACCTTGAAGGAGGCCTTTGACATTACCTACGCTGTGCATCGCA TGGAAGAGGAAGGAGCCAGGCGACGCAAGTACGGTAGAGGGACCCGGGGCTTCCTTAGCGAGCTTAACGAAGATG ACGAGATGGACGATGATGAGGACGATGACATCACCGATGAAAACGATGTTCGCAGGCACCCAGGACAGCAGCAGC AGCAACCACGTCACCCTCAAGGCCAAGCTCCGCAAAATACGTCGCTAATGGAAGGCCGCGCAGGACCAAGGGATC AAGGGAACCGAGAGTGGCATCACGCACAGTCGCAGGGAGTTGGGAGGACGAGCGGGCCGAGCGCTTCGGTTGCTA GAACGGGTGCGAACAGGGCGAGGGGACACAAAGGTCGACAGTTTGAACTGGATCTGGACGGTGCGACTCTATTGG AGAGGAGGCAGCGACGGGCAGCAGCAAAGAAGGATTATCCGGGTAGCCCATTGCGGATGGAAGGACTCAGGGTGG ACGATCGTCCATCTGTGACGGCCCTTTAGTGCGGTCGACTCTCTCCCTTCTCACACCTCTATTCAACACCCCTGT TCCATTCGCCCTTGCTATTGATATCCCCTGGTCCATTCACTGTTTACTAGTCTCCTTTCACTTTCCCTGTCTCCT TCACTGATGTCTGTAACATCTCCTGTATTTCCATATGAAGAAAGCACCTGGCATCACCCGAACGTTGTGTTGTAG AGGCACCTTGTATGTATTACTACTTTGATTGGTTCATTTAGCGTAGTAGTGTGTATTATTCCACCCACGTTCATG TACTGCAATGAGTTTCTCCCCTGGAACAATGGTATTTATTGTCATAATCCCAA |
| Length | 2228 |
Gene
| Sequence id | CopciAB_451611.T0 |
|---|---|
| Sequence |
>CopciAB_451611.T0 AATTCATCCTTTTTCGATCCCCCATCTCCACAATGCCTCCAACCGGCGTCGTCGATGCTTTCAAGAGTGTGTCGT CTTCATTGTTCTCGATGTCCAAACGCCCCTAATCACAGCGACCAGACCTCATTCGCCACGGCAAGCACCACAACC ACGTACAGCCGCCCAGACAGAACGAACAGGTGCCTTTGAATTCTCAGTATAATCTTCACATCCCAAGTATGCTCA CGTGCACCCAAGCCTCGTCAGCATCAGCCTCAAGGCCACAACGGCCCCGCCCCTGCCCAGGCGAGCAAACGCCAC AATGAGCAACCCACAGCACAGGCTGCTCAGCCCCACGAAAAGTCGCCACCCCCACGAGAAGCAGCTGAAATGATC GTCAACCAGGAACGCGCTGAAAAGTCAAAGATGCCAACTTACAAAGGCCTTGAGAGCTATACTTTGCTCGAGAAG ATGGGAGAGTGCGTCCTGCTCATCGTCGTTGCTGACGATAATCACCAGGCTGACATGATCCGTTAGTGGAGCATT CTCTAATGTCTACAAGGCTGTGGAGCACTCGACCAACCGCAAAGTTGCTGGTCAGTTACTCCCTGCATCGACCAA CTCTTTTGACTAAACCATCTCGCAAACAGTCAAGGTCGTCCGTAAATACGAACTCAATGCGTCACAGGTTCGTGG TTTTCCCTAACTTCTCAGTTTCCTCGTTTTCTTCCTTCCCAAACTAATCTTGATCTCTCCTTTGCCTGTTACTTG TCCTTGTTCATCTGCTGCTACACGCGGTCCTATCTATGCTTAGGCAGGGGAGAAACATCTCAATCCCCAATTCAA GAAGAAGCCGCGTGTGACAGAGGTGCCGTTCTCTTGCTGTTGACAAATGGGCCGTCTTACGATGTTCTGACCCTT TCTTTTCTAGCGGGCGAATATTCTCAAAGGTGTGTCTCTCGACGTGTTTCCACCTCCTATCTTACGCCGAATGCA GAGGTGCAGATCATGCGGGGCATTCAGCATCCCTCGATCGTGAAGCTTTACTCCTTTTCAGAATCGCCGGAGCAT TACTTCCTTGTGCTTGAGTGTAAGTGATGATCCCTCTTCCCATTGCAGACTCGGCTCATCCATGCGCTACAGTAA TGGAGGGTGGAGAACTATTCCATCAAATCGTCAAGCTCACCTACTTCAGTGAAAACCTCTCGCGTCATGTTATCC TCCAAGTCGCTCATGGCATACGACATCTCCATGAAGAACGGGGTGTCGTCCATCGCGATATCAAACCTGAAAACT TGCTATTCGAGCGCATAGACATCATCCCTTCCAAGAACCCAATAAAGCGACCGTACGATGAAGAGAAGGAAGACG AAGGCGAGTTCATACCCGGCGTTGGAGGAGGCGGTATTGGTCGCGTCAAGATCGCCGACTTTGGTCTTAGTAAGG TGGTCTGGGATGAGCAGACGATGACACCCTGTGGTACCGTTGGTTACACCGCACCGGAGATCGTCAAGGATGAGC GATACAGCAAGAGTGTAGATATGTGGGCTCTTGGCTGTGTCCTCTACACCCTTCTTTGTGGATTCCCTCCATTCT ACGATGAGAGTATCAATGTTCTCACTGAGAAGGTGGCACGGGGATACTATACCTTCCTCAGCCCTTGGTGGGACG ATATCAGCGAGAGCGGTACGCCAAGGTCAATTTATCTGTGTCCTATCCACTAACCCCTTTTCAGCAAAGGACCTC ATCAAGCATCTTCTTTGCGTCGATCCAGCCCAGAGGTATACTATCGACGAATTCCTCGCACACCCTTGGTGCAAC GCGGCGCCTGCTCCTCCGCCTCCCCCCACCCCTCTTGTTTACAGCGACAAATTCTCCATATCGGCGGCTGGCGGC CGACCTATTGATTCACCCCTTCTTTCCGCACCCCGGGGTAACTGGGAGGGTCGAAGTCCGGGTCTAGCCACCTTG AAGGAGGCCTTTGACATTACCTACGCTGTGCATCGCATGGAAGAGGAAGGAGCCAGGCGACGCAAGTACGGTAGA GGGACCCGGGGCTTCCTTAGCGAGCTTAACGAAGATGACGAGATGGACGATGATGAGGACGATGACATCACCGAT GAAAACGATGTTCGCAGGCACCCAGGACAGCAGCAGCAGCAACCACGTCACCCTCAAGGCCAAGCTCCGCAAAAT ACGTCGCTAATGGAAGGCCGCGCAGGACCAAGGGATCAAGGGAACCGAGAGTGGCATCACGCACAGTCGCAGGGA GTTGGGAGGACGAGCGGGCCGAGCGCTTCGGTTGCTAGAACGGGTGCGAACAGGGCGAGGGGACACAAAGGTCGA CAGTTTGAACTGGATCTGGACGGTGCGACTCTATTGGAGAGGAGGCAGCGACGGGCAGCAGCAAAGAAGGATTAT CCGGGTAGCCCATTGCGGATGGAAGGACTCAGGGTGGACGATCGTCCATCTGTGACGGCCCTTTAGTGCGGTCGA CTCTCTCCCTTCTCACACCTCTATTCAACACCCCTGTTCCATTCGCCCTTGCTATTGATATCCCCTGGTCCATTC ACTGTTTACTAGTCTCCTTTCACTTTCCCTGTCTCCTTCACTGATGTCTGTAACATCTCCTGTATTTCCATATGA AGAAAGCACCTGGCATCACCCGAACGTTGTGTTGTAGAGGCACCTTGTATGTATTACTACTTTGATTGGTTCATT TAGCGTAGTAGTGTGTATTATTCCACCCACGTTCATGTACTGCAATGAGTTTCTCCCCTGGAACAATGGTATTTA TTGTCATAATCCCAA |
| Length | 2790 |